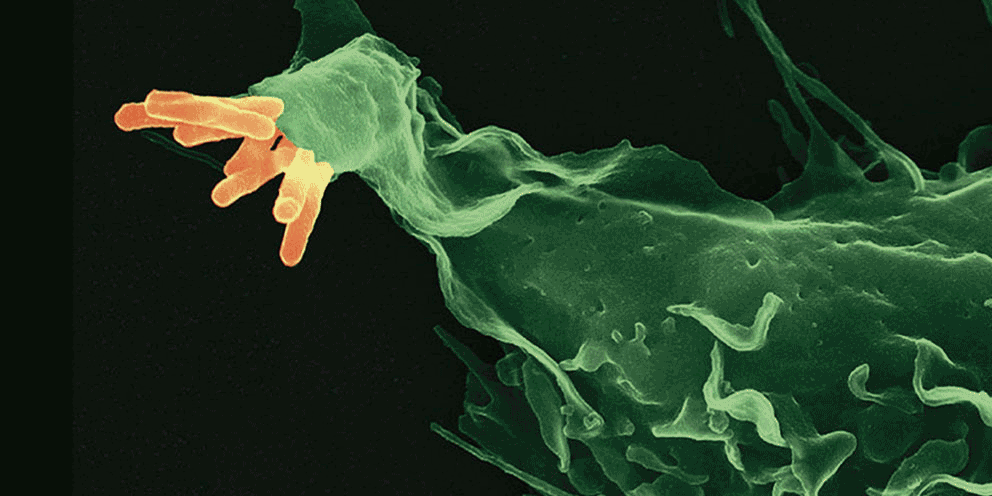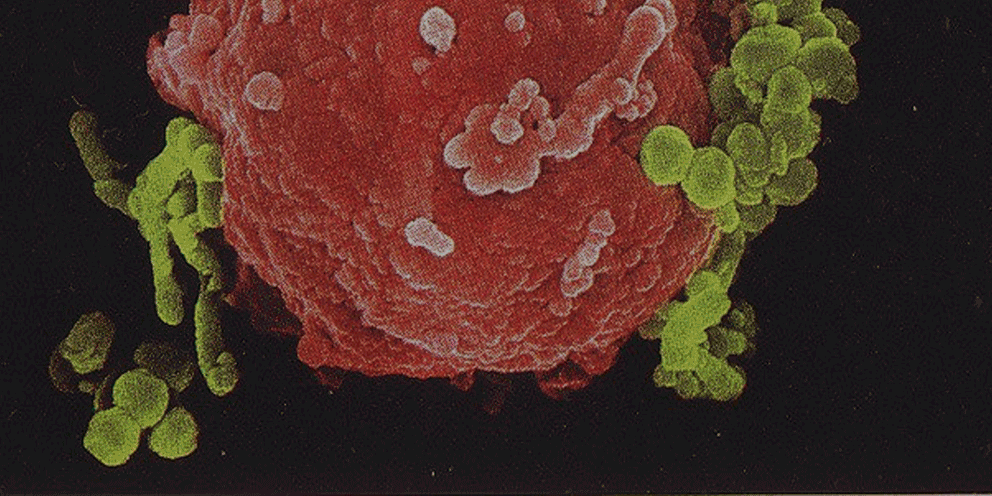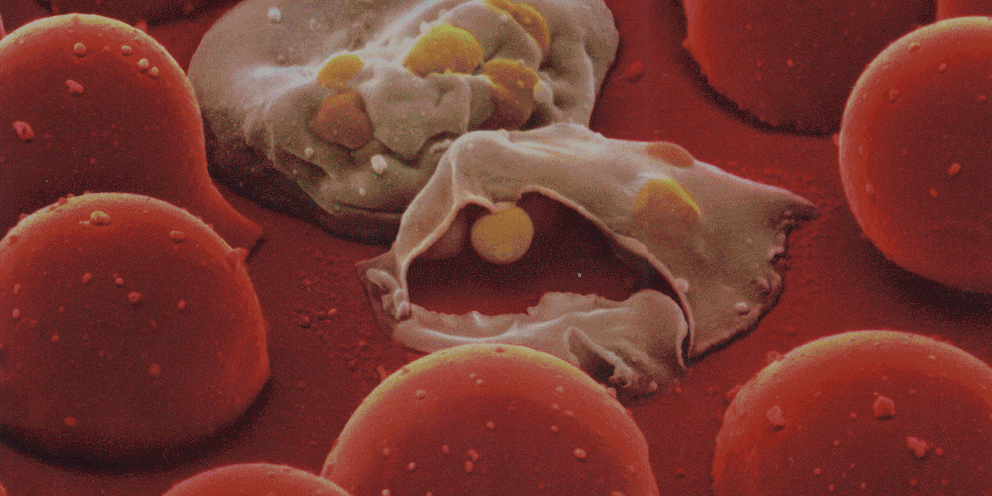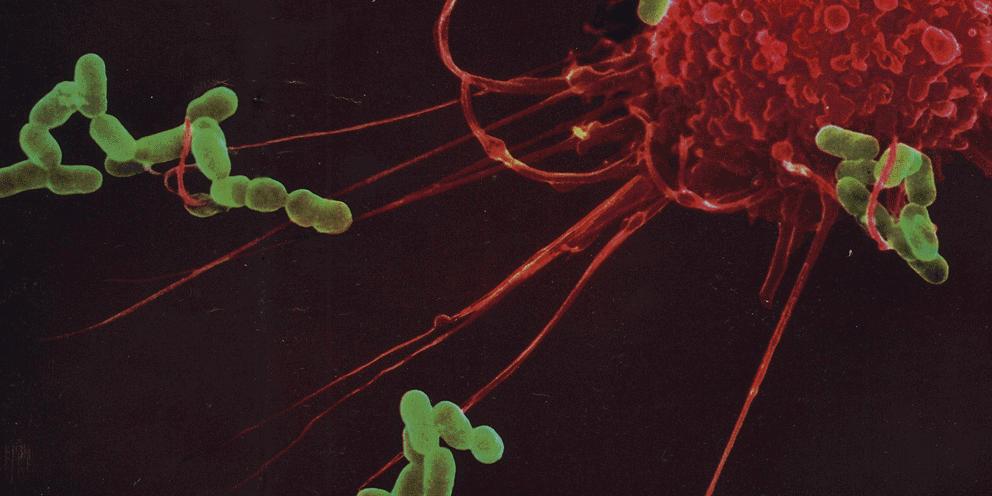
Several lines of evidence indicate that the fate of the infection process can be determined by just a few single individual encounters:
- Invasion of body surfaces by as little 10 pathogenic bacteria in some cases, can establish infection.
- From the site of infection, immune cells carry engulfed bacteria to deeper tissues, where they can disseminate to cause systemic infection.
- Infection progresses from several infectious loci resulting from clonal growth of an individual bacterium.
- Infection bottlenecks allows only very few individual bacteria to survive. These can then colonize tissues through spatial segregation of bacterial populations.
To date, one of the least-understood periods in the course of infectious disease lies between the initial inoculation with a dose of pathogens and the appearance of the first symptoms of disease, or the lack of appearance thereof. Application of novel single cell approaches to the study of infectious diseases can finally provide the much-needed resolution to analyze determinants of in vivo infection, individual cellular encounters, pathogen dissemination, formation of new infection foci, and infiltration of immune cells to form inflammatory lesions. Understanding this rich biology could significantly improve our ability to predict the outcomes of local host-pathogen encounters and overall disease progression, and enhance our ability to report on potential novel antimicrobial treatments.
Using a powerful combination of cutting-edge single cell genetic and genomic approaches, we wish to address what forms the basis for successful immune clearance, from the level of individual infected cells to that of the whole organism, and why, in some cases, sterilization is incomplete?