Our interest in the tumor microbiome was sparked almost by chance. In 2012 we published a paper (Straussman et al, Nature 2012) demonstrating that stromal cells can modulate the sensitivity of cancer cells to chemotherapy. In one of these cases, we later found that it was a mycoplasma bacteria infecting the stromal cells rather than the stromal cells themselves that conferred resistance to the drug gemcitabine. We followed up on this story demonstrating that the majority of human pancreatic tumors contain bacteria and that, in many cases, these bacteria can degrade the drug gemcitabine into an inactive metabolite. (Geller, Barzily-Rokni et al, Science 2017)
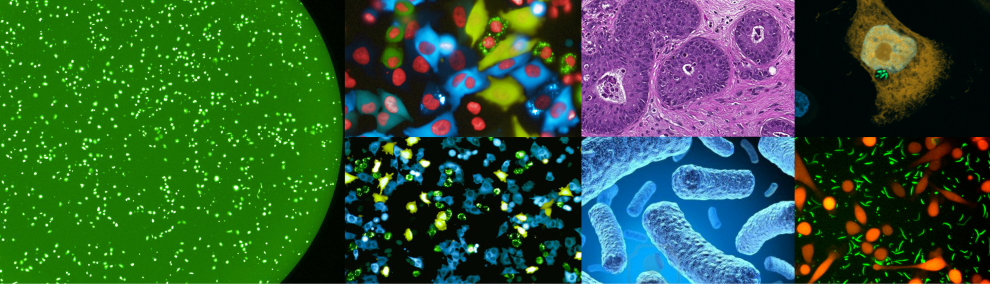
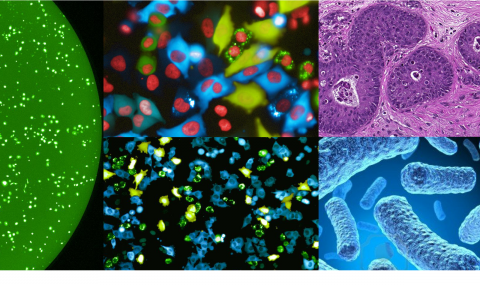
We then developed multiple methods to better detect, characterize and visualize bacteria in human tumors. We also expanded our study to many additional tumor types finding that each tumor type has a distinct microbiome composition. Visualization assays demonstrated that intra-tumor bacteria are mostly intracellular and are present in both cancer and immune cells. We also found interesting associations between the specific tumor bacteria (or their functions) and the makeup of the specific microenvironment these bacteria live in. (Nejman, Livyatan, Fuks et al, Science 2020).
The fact that bacteria are present inside tumor cells drove us to explore if peptides from these bacteria are present on the HLA of cancer cells, and may be detected by tumor infiltrating lymphocytes. In a collaboration with the lab of Dr. Yardena Samuels we indeed demonstrated that peptides derived from intracellular bacteria are presented on HLA-I and HLA-II molecules in melanoma tumors and can elicit immune reactivity (Kalaora, Nagler et al, Nature 2021).
In recent years, we also expanded our focus of the tumor microbiome to include fungi - the tumor mycobiome. In collaboration with the lab of Dr. Rob Knight, we studied fungi in 35 tumor types, using multiple methods including next generation sequencing, and staining of tumor sections. Fungi were detected in all tumor types and demonstrated tumor-type specific profiles. Similar to bacteria, fungi were visualized in both cancer and immune cells. We found interesting fungal-bacterial-immune cell associations. In addition, we found specific fungi to be associated with different clinical outcomes, such as response to immunotherapy and overall survival (Narunsky-Haziza, Sepich-Poore, Livyatan et al, Cell 2022).
We currently continue to study various aspects of the tumor microbiome. Some examples include:
- Characterization of the human tumor microbiome across additional tumor types and normal tissues.
- The effect of the tumor (and gut) microbiome on resistance to cytotoxic, targeted and immune mediated anti-cancer therapies.
- Characterization of the cross talk between bacteria and human cancer cells.
- Finding novel ways to manipulate the tumor/gut microbiome in order to affect different hallmarks of cancer.