Microstructure of the bacterial chromosome
One of the stunning surprises of the human genome project is that the number of genes in Homo sapiens, about 30,000, is not much larger than the number of genes (~4,500) needed to code for E. coli, a common bacterial cell in our guts. The bacterial genome, contained in a single DNA molecule, must be therefore long enough to carry all this information. In fact, with about 4.5 million base pairs, the genome's length is about 1 mm, a factor of 1000 larger than the diameter of our tiny E. coli cell. For it to fit inside, the genome must be compacted, and yet preserve its functions in cellular physiology. One of the important factors contributing to chromosome structure in bacteria is the association of DNA with a group of nucleoid-associated proteins such as IHF, HU, FIS and H-NS.
We study complexes formed between these proteins and single DNA molecules using single molecule techniques, in order to understand the architecture of the nucleoid. We probe the elasticity of twist-relaxed and supercoiled protein-DNA complexes, and the way these proteins deform DNA with fluorescence resonance energy transfer (FRET).
“DNA-protein interactions and bacterial chromosome architecture”
J. Stavans and A. B. Oppenheim, Phys. Biol. 3, R1 (2006).
IHF
Integration host factor or IHF assists in many processes involving high-order protein-DNA complexes such as in replication, regulation of transcription, and site-specific recombination, by binding at sequence-specific sites. At these latter, it bends DNA by about ~160o, the largest known among small proteins. What is then the reason for having more than 20,000 copies per cell if there are only of the order of 1000 known specific sites? Our elasticity studies have demonstrated that the non-specific interaction of this protein with DNA is substantial, bending the latter and producing more compact conformations. This effect, which is strongly salt-dependent, can contribute to the compact conformations of the nucleoid in stationary phase, during which the number of copies of IHF shoots up by a factor of 5!
"Compaction of Single DNA Molecules Induced by Binding of Integration Host Factor (IHF)"
B.M. Jaffar Ali, R. Amit, I. Braslavsky, A.B. Oppenheim, O. Gileadi and J. Stavans, Proc. Natl. Acad. Sci. USA 98, 10658 (2001).
H-NS
H-NS present in about 20,000 in the cell. It is a global regulator, controlling the expression of around 5% of genes. Most of these are related to the response of bacteria to environmental stimuli such as temperature, pH and osmolarity changes. Remarkably, this protein can downregulate gene expression from a high affinity site far away from a promoter (silencing), a phenomenon which has been demonstrated in the control of pathogenicity islands. Our experiments probing the elasticity of single H-NS-DNA complexes have shown that H-NS can rigidify DNA upon binding, a clear sign that it polymerizes along tracts of the DNA substrate. This polymerization is dependent upon temperature and osmolarity, rationalizing how this protein works as an environmental sensor.
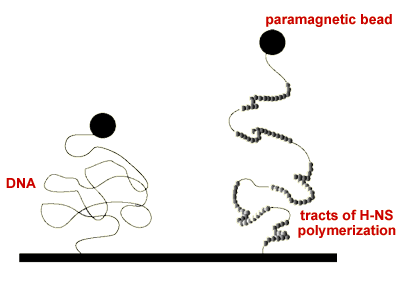
"Increased Bending Rigidity of Single DNA Molecules by H-NS, a Temperature and Osmolarity Sensor"
R. Amit, A. B. Oppenheim, and J. Stavans, Biophys. Jour. 84, 2467 (2003).
"Single Molecule Elasticity Measurements: a Biophysical Approach to Bacterial Nucleoid Organization"
R. Amit, A. B. Oppenheim, and J. Stavans, Biophys. Jour. 87, 1392 (2004).
HU
HU is a highly conserved protein from Mycoplasma one of the simplest cells, to extremophiles. In bacteria like E. coli, it is a highly abundant protein, being present in about 30,000 copies. HU does not recognize specific DNA sequences and binds homogeneously across the nucleoid, contributing to the maintenance of its supercoiled state.
Our FRET studies of short DNA molecules have shown that at small coverage, HU bends DNA by about 60o per dimer in a cooperative fashion. The highly bent conformations we observe by single-pair FRET suggest the existence of high-order complex formation, in which a number of HU dimers bend DNA forming a structure, the compactosome, reminiscent of eukaryotic nucleosomes. High protein/DNA ratios interfere with these structures, and lead to complexes with higher bending rigidity than bare DNA.
 Our studies of the elasticity of single HU-DNA complexes exhibit this non-monotonic behavior as well, exhibiting highly compact complexes at low HU conentrations and stiffer extended complexes at high HU concentrations. Remarkably, the elastic behavior of supercoiled complexes indicates that HU can lower the twist modulus of DNA by nearly a factor of ten. This effect may be behind the ability of HU to enable proper phasing in distal DNA segment interaction as in the Gal repressosome.
Our studies of the elasticity of single HU-DNA complexes exhibit this non-monotonic behavior as well, exhibiting highly compact complexes at low HU conentrations and stiffer extended complexes at high HU concentrations. Remarkably, the elastic behavior of supercoiled complexes indicates that HU can lower the twist modulus of DNA by nearly a factor of ten. This effect may be behind the ability of HU to enable proper phasing in distal DNA segment interaction as in the Gal repressosome.
"Fluorescence Resonance Energy Transfer Evidence for High-Order Macromolecular Structure of HU-DNA Complexes"
D. Sagi, N. Friedman, C. Vorgias,A. B. Oppenheim and J. Stavans, Jour. Mol. Biol. 341/2 , 419 (2004).


