Bacteria are remarkably skilled at doing more with less, utilizing clever regulatory mechanisms to increase their adaptability despite limited genomic realty. Our lab studies cis-acting riboregulators: 5’UTR-encoded RNA elements that sense intracellular metabolites and in response turn gene expression “ON” or “OFF.” This mode of regulation allows an mRNA to autonomously regulate its own expression as a function of metabolite concentration via premature transcription termination or by modulating its translation efficiency. Riboregulators present promising targets for therapeutics due to their vital roles in various metabolic nodes, bacterial virulence, and antibiotic resistance. In addition, their plug-and-play properties and high ligand specificities have highlighted these RNA-biosensors for diverse synthetic biology applications.
Our lab applies (meta-)transcriptomics and functional genomics approaches to study RNA biosensors across bacteria, with the goal of developing new tools for synthetic biology and environmental monitoring.
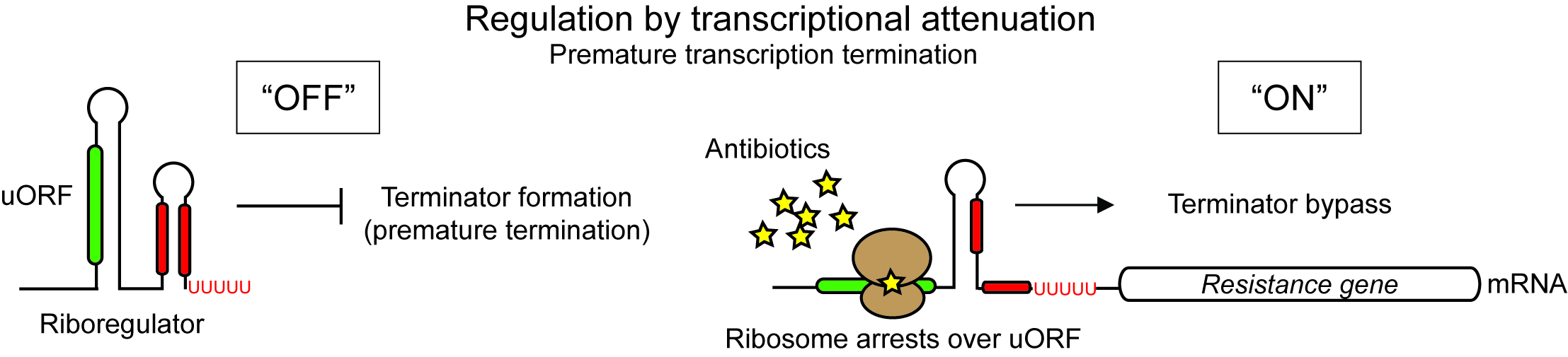
Attenuator riboregulators can use ribosome inhibition as a proxy for measuring antibiotic concentrations. Ribosome inhibition induces structural changes in the regulatory RNA and turns the expression of antibiotic resistance genes "ON".
Relevant publications:
- Dar D et al. Term-seq reveals abundant riboregulation of antibiotics resistance in bacteria. Science (2016).
- Dar D, Sorek R. Regulation of antibiotic-resistance by non-coding RNAs in bacteria. Current Opinion in Microbiology (2017).