Primers-4-Yeast
Primers-4-Yeast allows users to design primers (single or batch) for gene targeting of PCR-based transformation cassettes into S. cerevisiae, and for validation of correct insertion.
Use the following links to jump to page sections:
|
Targeting primers |
Verification primers |
Other |
|---|---|---|
|
|
||
ORF or gene name list Input
Paste a list of target Gene names and/or Open Reading Frames (ORFs) separated by line breaks (<Enter>),or use the search field on the right to find and insert entries one-by-one.
The list can include a mixture of ORF and gene names. Duplicates will be removed from results.
Gene name is the standard name, e.g. TFC3. ORF is the systematic name, e.g. YAL001C.
Primers-4-Yeast was designed using the S288C reference genome sequence. Researchers working on other S. cerevisiae backgrounds (e.g. W303) should be aware of the possibility of divergence of the target sequence from S288C, and are encouraged to verify that the targeting sequences in the primers match in the different background, as mis-matches can cause low transformation efficiencies.
Please notice that while the size of the gene list is not limited, inputs of large sets (>500 entries) will result in long processing time of up to a few minutes. Users that require very large sets of primers are welcome to contact us.
Targeting primers
Knockout (deletion)
Knockout (KO) primers are intended to amplify a selection cassette from a plasmid, with flanking sequences directing it to recombine in place of the target gene.

Forward primer: 40 base pairs (bp) upstream to the gene’s start codon (ATG) (including the ATG in the primer), followed by the “forward primer” sequence of the transformation cassette (plasmid dependent). Note that the genomic targeting sequence of this primer is the same as for N'-tagging forward primer.
Reverse primer: The reverse complement of 40bp downstream to the gene’s Stop-codon (including the Stop-codon in the primer), followed by the “reverse primer” sequence of the transformation cassette (plasmid dependent). Note that the genomic targeting sequence of this primer is the same as for C'-tagging reverse primer.
To verify correct insertion use WT Check primers, or 5UTR/3UTR Check primers.
DAmP (Decreased Abundance by mRNA Perturbation)
DAmP primers are intended to amplify a selection cassette from a plasmid, with flanking sequences directing it to recombine in place of the gene’s 3’UTR (immediately after the stop codon). The presence of the selection marker promoter which replaces the original gene’s 3’UTR perturbs gene expression, as described by Schuldiner et al. 2005 (References).
Note that it is recommended to perform the DAmP switch in diploid cells if the target gene is essential. Following sporulation the haploids have less suppressor mutations.

Forward primer: 40bp upstream to the gene’s Stop-codon (including the Stop-codon in the primer), followed by the “forward primer” sequence of the transformation cassette (plasmid dependent).
Reverse primer: The reverse complement of 40bp after the gene’s Stop-codon (excluding the Stop-codon from the primer), followed by the “reverse primer” sequence of the transformation cassette (plasmid dependent). Note that the genomic targeting sequence of this primer is the same as for C'-tagging reverse primer.
To verify correct insertion use C’-tag Check primers.
C’-tagging
C’ tagging primers are intended to amplify a cassette containing a tag to be fused in frame to the desired protein at its C terminus, followed by a generic terminator and a selection marker. Primers contain flanking sequences directing the tagging cassette to recombine into the gene end, creating a fused protein.
Templates used for C’ tags should encode a stop codon at the end of the tag’s sequence, and a terminator for transcription termination before the selection cassette.

Forward primer: 40bp before the gene’s Stop-codon (excluding the Stop-codon from the primer), followed by the “forward primer” sequence of the transformation cassette (plasmid dependent).
Reverse primer: The reverse complement of 40bp downstream to the gene’s Stop-codon (including the Stop-codon in the primer), followed by the “reverse primer” sequence of the transformation cassette (plasmid dependent). Note that the genomic targeting sequence of this primer is the same as for KO reverse primer.
To verify correct insertion use C’-tag Check primers.
N’-tagging
N’ tagging primers are intended to amplify a cassette containing a selection marker followed by a promoter and a tag to be fused to the desired protein at its N terminus. Primers contain flanking sequences directing the tagging cassette to recombine into the gene start, creating a fused protein.
Templates used for N’ tags should contain a selection marker followed by a promoter for transcription, and a start codon for the tag to ensure initiation of translation of the fused protein.

Forward primer: 40bp upstream to the gene’s start codon (ATG) (including the ATG in the primer), followed by the “forward primer” sequence of the transformation cassette (plasmid dependent). Note that the genomic targeting sequence of this primer is the same as for KO forward primer.
Reverse primer: The reverse complement of 40bp after the gene’s start codon (ATG) (excluding the ATG from the primer), followed by the “reverse primer” sequence of the transformation cassette (plasmid dependent).
To verify correct insertion use N’-tag Check primers.
Please note that since targeting primers are dictated by the gene and plasmid sequences, and in addition are longer than normal primers, they have the potential to cause a failure of the PCR reaction. Although failure could be due to strong secondary structures, differences in melting temperatures and other factors, we find these cases to be rare.
Plasmid sets
Several sets of plasmids encoding transformation cassettes have been published and are widely used for S. cerevisiae homologous recombination (References). Primers-4-Yeast contains primers designed for PCR amplification using these common plasmids. Each plasmid set uses common sequences for amplification of the cassettes:
| Plasmid set | Name | Sequence | Orientation | Targeting type |
|---|---|---|---|---|
| pFA6 | F1* | cggatccccgggttaattaa | F | KO |
| R1** | gaattcgagctcgtttaaac | R | KO / C' tagging / DAmP | |
| F2* | cggatccccgggttaattaa | F | C' tagging / DAmP | |
| F4** | gaattcgagctcgtttaaac | F | N' tagging | |
| R2 | cattttgagatccgggtttt | R | N' tagging - (no tag) | |
| R3 | gcactgagcagcgtaatctg | R | N' tagging - with 3HA tagging | |
| R4 | acgcggaaccagatccgatt | R | N' tagging - with GST tagging | |
| R5 | tttgtatagttcatccatgc | R | N' tagging - with GFP(S65T) tagging | |
|
* F1 and F2 are identical in sequence. ** R1 and F4 are identical in sequence. |
||||
| pYM | S1 | cgtacgctgcaggtcgac | F | KO / N' tagging |
| S2 | atcgatgaattcgagctcg | R | KO / C' tagging | |
| S3 | cgtacgctgcaggtcgac | F | C' tagging | |
| S4 | catcgatgaattctctgtcg | R | N' tagging | |
|
DAmP § |
DAmP F | acatggaggcccagaataccc | F | DAmP |
| DAmP R | cagtatagcgaccagcattcac | R | DAmP | |
| pCg / pKl | F | cacaggaaacagctatgacc | F | KO |
| R | gttgtaaaacgacggccagt | R | KO | |
§The DAmP library was built using a pFA6-KAN construct, although the primers that were used were not the pFA6 generic amplification sequences (F1 & R1), rather new sequences found internaly (in respect to the F1&R1), that we refer to as DAmP F&R. Therefore you may choose to perform DAmP in this way, or to use the generic pFA6 or pYM plasmid sequences to amplify the selection cassette.
If you are planning to publish a new plasmid set and would like us to include it in Primers-4-Yeast please contact us.
Selection markers
Selection markers contain either a gene conferring antibiotic resistance (e.g KAN, NAT) or a complementation of an auxotrophic mutation (e.g HIS3, URA3, MET15 etc.), and a promoter and terminator for constitutive expression. Examples for commonly used promoters and terminators: TEF, ADH1, CYC1, PGK etc.
Design primers for a custom plasmid
If a custom plasmid (not represented in Primers-4-Yeast) is required, users should input the plasmid name and cassette amplification sequences (forward and reverse primer sequences), and select the gene targeting types. Primers-4-Yeast will design primers for amplification of the cassette from the custom plasmid, and their insertion into each ORF from the input list, based on the targeting type (see above).
Check primers
Check primers are intended for verification of correct insertion of the transformation cassettes to the desired location (gene). Primers were generated using BatchPrimer3 and are on average 20bp long, have 40% GC content, and contain a one bp GC clamp (at least one G or C at the 3’ end). Primers have an average melting temperature of 60°C, and we recommend setting this as the annealing temperature, as we successfully tested 129 primers so far with this parameter using various polymerases. Please contact us if you experiece problems with specific primers.
Note – following transformations yeast often become aneuploid and so the mere presence of a selection cassette in your gene’s locus does not verify that all copies of your gene have been tagged or deleted. Therefore it is recommended to use several PCR and phenotypic verification strategies (such as “WT check” below) for gene deletions and tagging.
WT check
Wild-type (WT) check primers are intended to test for the presence of a copy of the gene in the yeast genome, and can therefore verify gene deletions. Notice that since a strain with the deletion in question would yield no bands in such a PCR reaction, positive controls that verify the template (using another primer pair that would yield a band) and the primers (a WT strain, which is not deleted for the gene in question) are necessary.

Forward and reverse primer pairs were designed to amplify a fragment from within the coding sequence of the gene, creating a 350bp amplicon on average. The forward primer is taken from the gene sense orientation(  ), and the reverse primer is taken from the anti-sense orientation (
), and the reverse primer is taken from the anti-sense orientation (  ).
).
5’UTR check
5’UTR check primers are intended to test the correct deletion of a gene or the insertion of a tag into the 5’ end of the gene in question (N’ tagging). For each gene, the forward primer is from the gene sense orientation, on average 400bp upstream to the start-codon (  ).
).

As the reverse primer for this PCR one should use a primer which is the reverse complement of the 5’ end of the selection marker (  ).
).
3’UTR check
3’UTR check primers are intended to test the correct deletion of a gene or the insertion of a tag into the 3’ end of the gene in question. For each gene, the reverse primer is from the gene anti-sense orientation (reverse), on average 400bp downstream of the stop codon (  ).
).

As the forward primer for this PCR one should use a primer from the 3’ end of the selection marker (  ).
).
C’-tag check
C’-tag check primers are intended to test the correct insertion of a tag (e.g. GFP tag) into the 3’ end of the gene in question. For each gene, the forward primer is from the gene’s coding strand, on average 400bp upstream to the stop-codon (  ).
).
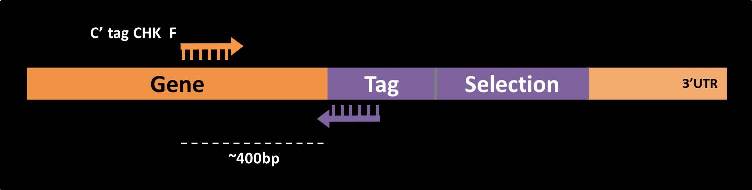
As the reverse primer for this PCR one should use a primer which is the reverse complement of the 5’ end of the tag (  ), such as the following:
), such as the following:
| Plasmid set | Name | Sequence |
|---|---|---|
| pFA6 |
F2 reverse complement |
ttaattaacccggggatccg |
| pYM |
S3 reverse complement |
gtcgacctgcagcgtacg |
N’-tag check
N’-tag check primers are intended to test the correct insertion of a tag or a promoter swap into the 5’ end of the gene in question. For each gene, the reverse primer is from the gene anti-sense strand (reverse), on average 400bp downstream of the start codon (  ).
).

As the forward primer for this PCR one should use a primer from the 3’ end of the tag (  ), such as the following:
), such as the following:
| Plasmid set | Name | Sequence |
|---|---|---|
| pFA6 |
R2 reverse complement |
aaaacccggatctcaaaatg |
|
R3 reverse complement |
cagattacgctgctcagtgc |
|
|
R4 reverse complement |
aatcggatctggttccgcgt |
|
|
R5 reverse complement |
gcatggatgaactatacaaa |
|
|
pYM |
S4 reverse complement |
cgacagagaattcatcgatg |
Results
A submitted query output will include a table of the designed primers, with each primer’s ORF, gene name, primer name, and primer sequence.
Results may be downloaded in the form of a comma separated values (.csv) file with additional primer information: length, orientation, description, primer pair, product size (for check primers), and the distances from ORF start and stop in base pairs.
