Research
The main goal of our group is to decipher the first principles of cellular self-organization processes such as protein folding, protein-protein assembly, and protein-DNA recognition. We are focused on advancing our understanding of biomolecules (the sequence-structure-function problem) using a battery of computational and theoretical methods that capture their chemical and physical nature as well as the billions of years of evolutional design.
We are focused on advancing our understanding of biomolecules (the sequence-structure-function problem) using a battery of computational and theoretical methods that capture their chemical and physical nature as well as the billions of years of evolutional design. Our main aim is to decipher the complexity of proteins and nucleic acids that results in self-recognition and cellular communication and therefore a biological function. Unraveling mysteries on the structure, assembly, and interactions at the molecular level have long-term implications for combating medical conditions such as Creuzfeld-Jacob (Mad Cow disease), Alzheimer, and cancer.
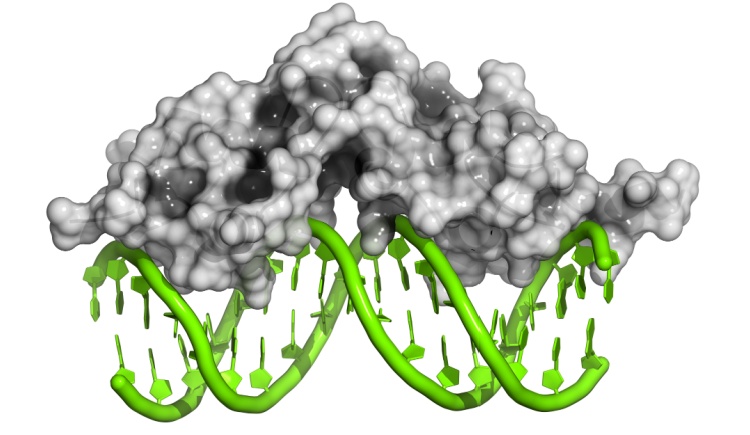
We are interested in studying fundamental questions of the mechanisms of protein folding and to formulate the forces that bias an efficient folding. In addition we focus on understanding mechanisms of protein association and the degree of protein plasticity involved in these reactions. We are also investing our efforts in deciphering the mechanisms and kinetics of DNA recognition by monomeric and multimeric proteins with the ultimate goal of understanding cellular communication from physical and molecular viewpoints using theoretical and computational tools, using structure based modeling.
News

Congratulations for Yoav (19/8/25) and Simona (2/11/25) defending their MSc thesis!