Legacy Research: Computational discovery of novel natural antibiotics
The need for new antibiotics
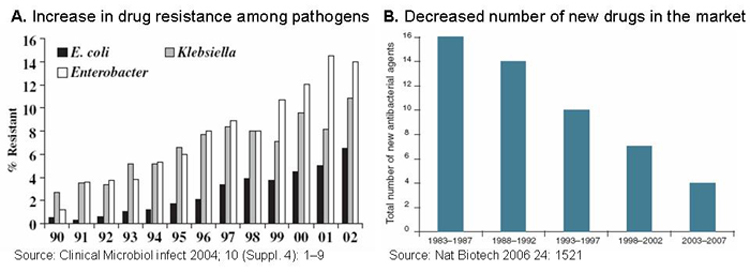
There is a growing need for new antimicrobial agents in the clinic. Drug resistance is spreading among pathogens, but the number of new antibiotics in the market is constantly decreasing. As a result, hospital-acquired bacterial infections now affect 1.7 million patients annually in the U.S. alone, and are responsible for 99,000 deaths every year. There is, therefore, a clear and urgent need to expand our clinical arsenal of antimicrobial drugs.
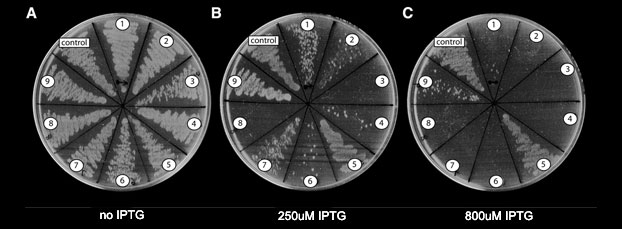
Large-scale discovery of genes toxic to bacteria
As part of our research we developed a computational method that detects, among a large number of microbial genomes, genes producing products that are toxic to bacteria. These genes were tested for their antimicrobial activity. Our approach was based on a by-product of the genome sequencing process, where initial assemblies invariably contain gaps due to DNA fragments that cannot be cloned in bacteria (Fig 1). We discovered that these uncloneable gaps are caused by genes that are toxic to E. coli (Sorek et al., Science 2007). Thousands of such genes from hundreds of bacteria were collected into the PanDaTox database. Among these toxic genes we were able to discover new types of microbial toxins, new restrictions enzymes, toxic small RNAs and toxic DNA binding motifs (Kimelman et al, 2012). Toxin-antitoxin systems, some of them form bacterial defense systems, are also discovered (Sberro et al, Molecular Cell 2013). We also studied antibiotics resistance and recently discovered new RNA-mediated mechanisms for regulation of antibiotic-resistance genes in the human microbiome (Dar et al, Science 2016).