Intercellular interactions during T cell activation and differentiation
Cells of the immune system are not acting autonomously; rather their responses strongly rely on signals that they receive from other cells in the system. Communication between immune cells is necessary to control immune responses in time and space, and probably takes an important part in immune recognition and information processing as well. The complexity of the intercellular communication network obscures analysis and understanding of its function. A possible approach towards simplification of the description of the system is by recognizing and studying recurrent modules, which may serve as simpler building blocks of the entire network. Our research focuses on the study of simple building blocks of the immune intercellular communication network, combining experimental approaches and mathematical modeling.
Read more
Studying the dynamics of T cell activation and differentiation using live cell imaging in microwells
Methods that allow for monitoring of individual cells over time, using live cell imaging, are essential for studying dynamical cellular processes in heterogeneous cell populations such as T lymphocytes. However, applying these methods to study activation and differentiation of primary T cells is challenging, due to their non-adherent nature, and their high motility upon activation through their antigen receptor. We developed a microfluidics based platform which enables the capture and long term culture of single primary T lymphocytes with minimal perturbation. We use this microwell array system to study dynamical processes and intercellular interactions within heterogeneous populations of primary T cells at the single cell level.
Read more
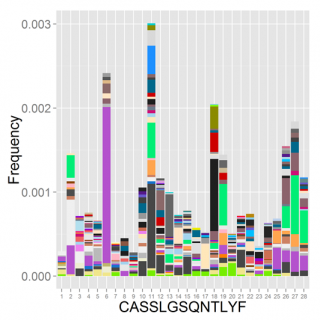
Characterizing T cell receptor repertoires using high throughput sequencing (TCR-seq)
Recognition of antigens by T cells relies on a diverse repertoire of T-cell receptor (TCR) chains. In the era of whole genome sequencing, the expressed repertoire of lymphocyte receptors remains “terra incognita” of unmapped genetic information that is highly variable between individuals and also dynamically changing throughout life. Global characterization of these repertoires using recent advances in high-throughput sequencing is expected to revolutionize our understanding of the adaptive immune system and its related pathologies, ranging from immunodeficiencies to cancer and autoimmunity. We developed a methodology for using high throughput sequencing to map TCR repertoires at high resolution (TCR-seq). We apply TCR-seq to study various immune perturbations, including repertoire changes in response to infection, vaccination or in autoimmunity.
Read more