Signaling pathways control many of the hallmarks of cancer such as aberrant cell growth, fate, survival, and genome maintenance. Adaptive pathway rewiring has an important role in innate, upfront resistance to targeted therapies. Interestingly, all of the known cancer driver genes can be classified into a dozen or so core signaling pathways which we can exploit to better understand pathway rewiring
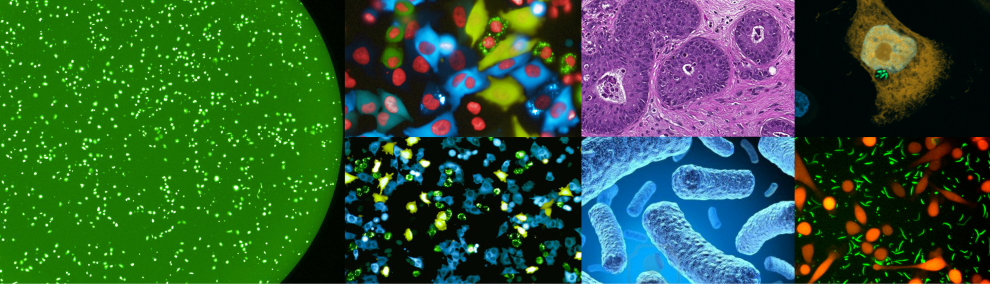
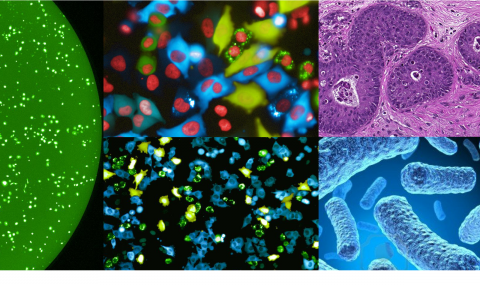
Current methods aimed at profiling pathway activity can either look at the population level (e.g. westerns, qPCR), or at the single cell level using low-throughput methods to study live or fixed cells (e.g. GFP based reporters, IHC).
Our lab has developed a new platform that allows us to trace the activity of multiple signaling pathways at the single cell level over time and in a high-throughput manner. For example, from each well in a 384-well plate we are able to follow the activity of 12-16 different signaling pathways over time and in a single cell resolution.
Using our ‘signalome’ system, we now follow multiple biological questions, including adaptive pathway rewiring in response to anti-cancer treatments.