Publications
2025
-
(2025) Science Advances. 11, 49, eadu0315. Abstract
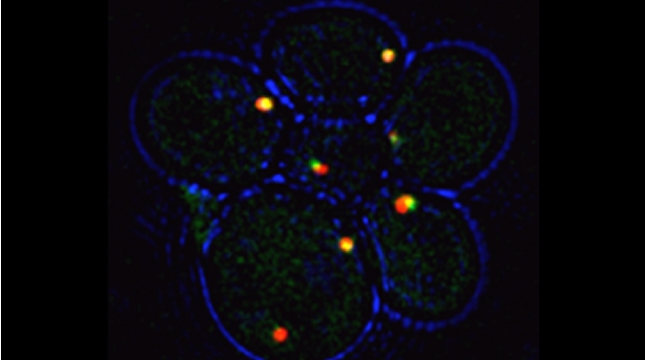
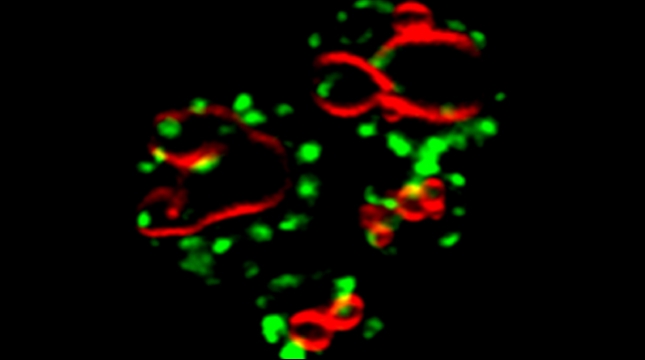
Prokaryotes use polycistronic transcription (operons) to express multiple messenger RNAs (mRNAs) from a single promoter to coexpress functionally related genes. However, how do eukaryotes, which express monocistronic messages, achieve the same regulation? Previously, we demonstrated that yeast uses RNA operons, i.e., mRNAs assembled in trans (transperons), to control multiple cellular pathways such as the heat shock response (HSR). As the HSR is conserved from yeast to mammals, we used single-molecule RNA labeling and pulldown techniques to demonstrate that mammalian heat shock protein (HSP) mRNAs also form operons upon transcription during heat stress. HSP RNA operon formation is dependent on the heat shock factor 1 transcription factor and intra- and interchromosomal interactions between the HSP genes. Work in yeast identified a conserved RNA sequence motif and histone H4 functions that act downstream thereof to regulate transperon assembly. Our work highlights the evolutionarily conserved regulation of the HSR and for RNA operons in eukaryotic gene regulation.
-
(2025) Communications Biology. 8, 242. Abstract
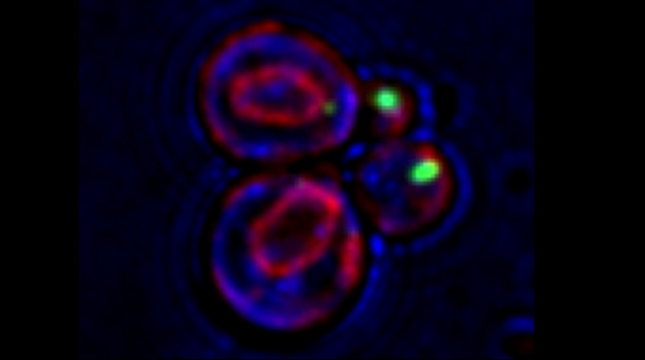
Analysis of single-molecule fluorescent in situ hybridization (smFISH) images is important to translate cellular image data into a quantifiable format. Although smFISH is the gold standard for RNA localization measurements, there are no freely available, user-friendly applications for assaying messenger RNA (mRNA) localization to organelles. EASI-ORC (Efficient Analysis and Segmentation of smFISH Images for Organelle-RNA Colocalization) is a novel pipeline for the automated analysis of multiple smFISH images of yeast cells. EASI-ORC automates the segmentation of cells and organelles, identifies bona fide smFISH signals, and measures mRNA-organelle colocalization. EASI-ORC is efficient, unbiased, and plots the colocalization data and statistical analyses. EASI-ORC utilizes existing ImageJ plugins and original scripts, thus allowing for free access and ease-of-use. To circumvent technical literacy issues, a step-by-step user guide is provided. EASI-ORC offers a robust solution to smFISH image analysis - one that saves time, effort and provides consistent measurements of mRNA-organelle colocalization in yeast. (Figure presented.)
2023
-
(2023) eLife. 12, e83584. Abstract
Full-length mRNAs transfer between adjacent mammalian cells via direct cell-to-cell connections called tunneling nanotubes (TNTs). However, the extent of mRNA transfer at the transcriptome-wide level (the 'transferome') is unknown. Here, we analyzed the transferome in an in vitro human-mouse cell co-culture model using RNA-sequencing. We found that mRNA transfer is non-selective, prevalent across the human transcriptome, and that the amount of transfer to mouse embryonic fibroblasts (MEFs) strongly correlates with the endogenous level of gene expression in donor human breast cancer cells. Typically,
2022
-
(2022) Trends in Genetics. 38, 12, p. 1217-1227 Abstract
Coordinated gene expression allows spatiotemporal control of cellular processes and is achieved by the cotranscription/translation of functionally related genes/proteins. Prokaryotes evolved polycistronic messages (operons) to confer expression from a single promoter to efficiently cotranslate proteins functioning on the same pathway. Yet, despite having far greater diversity (e.g., gene number, distribution, modes of expression), eukaryotic cells employ individual promoters and monocistronic messages. Although gene expression is modular, it does not account for how eukaryotes achieve coordinated localized translation. The RNA operon theory states that mRNAs derived from different chromosomes assemble into ribonucleoprotein particles (RNPs) that act as functional operons to generate protein cohorts upon cotranslation. Work in yeast has now validated this theory and shown that intergenic associations and noncanonical histone functions create pathway-specific RNA operons (transperons) that regulate cell physiology. Herein the involvement of chromatin organization in transperon formation and programmed gene coexpression is discussed.
-
(2022) Bio-Protocol. 12, 1, e4274. Abstract
RNA-RNA and RNA-protein interactions are involved in the regulation of gene expression. Here, we describe an updated and extended version of our RNA purification and protein identification (RaPID) protocol for the pulldown of aptamer-tagged mRNAs by affinity purification. The method takes advantage of the high affinity interaction between the MS2 RNA aptamer and the MS2 coat protein (MCP), as well as that between streptavidin-binding peptide (SBP) and streptavidin. Thus, it employs MCP-SBP fusions to affinity purify MS2-tagged target RNAs of interest over immobilized streptavidin. Purified aptamer-tagged mRNAs, along with any associated RNAs and proteins, are then sent for RNA sequencing (RaPID-seq) or mass spectrometry (RaPID-MS), which allows for the identification of bound cohort RNAs and proteins, respectively.
2021
-
(2021) eLife. 10, e66050. Abstract
Prokaryotes utilize polycistronic messages (operons) to co-translate proteins involved in the same biological processes. Whether eukaryotes achieve similar regulation by selectively assembling and translating monocistronic messages derived from different chromosomes is unknown. We employed transcript-specific RNA pulldowns and RNA-seq/RT-PCR to identify yeast mRNAs that co-precipitate as ribonucleoprotein (RNP) complexes. Consistent with the hypothesis of eukaryotic RNA operons, mRNAs encoding components of the mating pathway, heat shock proteins, and mitochondrial outer membrane proteins multiplex in trans, forming discrete messenger ribonucleoprotein (mRNP) complexes (called transperons). Chromatin capture and allele tagging experiments reveal that genes encoding multiplexed mRNAs physically interact; thus, RNA assembly may result from co-regulated gene expression. Transperon assembly and function depends upon histone H4, and its depletion leads to defects in RNA multiplexing, decreased pheromone responsiveness and mating, and increased heat shock sensitivity. We propose that intergenic associations and non-canonical histone H4 functions contribute to transperon formation in eukaryotic cells and regulate cell physiology.
-
(2021) Biochemical Society Transactions. 49, 1, p. 145-160 Abstract
It was already suggested in the early 70s that RNA molecules might transfer between mammalian cells in culture. Yet, more direct evidence for RNA transfer in animal and plant cells was only provided decades later, as this field became established. In this mini-review, we will describe evidence for the transfer of different types of RNA between cells through tunneling nanotubes (TNTs). TNTs are long, yet thin, open-ended cellular protrusions that are structurally distinct from filopodia. TNTs connect cells and can transfer many types of cargo, including small molecules, proteins, vesicles, pathogens, and organelles. Recent work has shown that TNTs can also transfer mRNAs, viral RNAs and non-coding RNAs. Here, we will review the evidence for TNT-mediated RNA transfer, discuss the technical challenges in this field, and conjecture about the possible significance of this pathway in health and disease.
2020
-
(2020) RNA Tagging. Heinlein M.(eds.). p. 195-214 Abstract
Intercellular communication is a major hallmark of multicellular organisms and is responsible for coordinating cell and tissue differentiation, immune responses, synaptic transmission, and both paracrine and endocrine signaling, for example. Small molecules, peptides, and proteins have all been studied extensively as mediators of intercellular communication; however, RNAs have also been shown recently to transfer between cells. In mammalian cells, microRNAs, tRNAs, short noncoding RNAs, mRNA fragments, as well as full-length mRNAs have all been shown to transfer between cells either by exosomes or by membrane nanotubes. We have previously described nanotube-mediated cell-cell transfer of specific mRNAs between heterologous mammalian cell types cultured in vitro. Here, we describe a simple method for the unbiased and quantitative identification of the complete range of transferred mRNAs (i.e., the mRNA transferome) in one population of mammalian cells following co-culture with another population. After co-culture, the individual cell populations are sorted by magnetic bead-mediated cell sorting and the transferred RNAs are then identified by downstream analysis methods, such as RNA sequencing. Application of this technique not only allows for determination of the mRNA transferome, but can also reveal changes in the native transcriptome of a cell population after co-culture. This can indicate the effect that co-culture and intercellular transfer of mRNA have upon cell physiology.
2019
-
(2019) Imaging Gene Expression. Shav-Tal Y.(eds.). New York, NY: . p. 109-129 (trueMethods in Molecular Biology). Abstract
In eukaryotic cells, a small percentage of mRNA molecules can undergo transfer from one cell to another. mRNA transfer occurs primarily via membrane nanotubes, which are long thin protrusions that are produced by numerous cell types and can connect cells that can be up to hundreds of microns apart. Potentially, mRNAs might also transfer via extracellular vesicles (EVs). Here we describe a method to detect transferred mRNA in cocultures of two different cell types and to distinguish between nanotube- and EVs-mediated transfer. This method uses single molecule fluorescent in situ hybridization (smFISH) to provide an accurate and quantitative detection of transferred mRNA molecules and their subcellular localization. Following the guidelines presented here will allow the user to investigate mRNA transfer of most transcripts in any co-culture system. In addition, we present modifications that improve nanotube preservation during the smFISH procedure.
-
(2019) PLoS Genetics. 15, 7, e1008248. Abstract
The localization of mRNAs encoding secreted/membrane proteins (mSMPs) to the endoplasmic reticulum (ER) likely facilitates the co-translational translocation of secreted proteins. However, studies have shown that mSMP recruitment to the ER in eukaryotes can occur in a manner that is independent of the ribosome, translational control, and the signal recognition particle, although the mechanism remains largely unknown. Here, we identify a cis-acting RNA sequence motif that enhances mSMP localization to the ER and appears to increase mRNA stability, and both the synthesis and secretion of secretome proteins. Termed SECReTE, for secretion-enhancing cis regulatory targeting element, this motif is enriched in mRNAs encoding secretome proteins translated on the ER in eukaryotes and on the inner membrane of prokaryotes. SECReTE consists of >= 10 nucleotide triplet repeats enriched with pyrimidine (C/U) every third base (i.e. NNY, where N = any nucleotide, Y = pyrimidine) and can be present in the untranslated as well as the coding regions of the mRNA. Synonymous mutations that elevate the SECReTE count in a given mRNA (e.g. SUC2, HSP150, and CCW12) lead to an increase in protein secretion in yeast, while a reduction in count led to less secretion and physiological defects. Moreover, the addition of SECReTE to the 3'UTR of an mRNA for an exogenously expressed protein (e.g. GFP) led to its increased secretion from yeast cells. Thus, SECReTE constitutes a novel RNA motif that facilitates ER-localized mRNA translation and protein secretion.
-
(2019) PLoS Biology. 17, 3, e3000182. Abstract
In experimental evolution, scientists evolve organisms in the lab, typically by challenging them to new environmental conditions. How best to evolve a desired trait? Should the challenge be applied abruptly, gradually, periodically, sporadically? Should one apply chemical mutagenesis, and do strains with high innate mutation rate evolve faster? What are ideal population sizes of evolving populations? There are endless strategies, beyond those that can be exposed by individual labs. We therefore arranged a community challenge, Evolthon, in which students and scientists from different labs were asked to evolve Escherichia coli or Saccharomyces cerevisiae for an abiotic stresslow temperature. About 30 participants from around the world explored diverse environmental and genetic regimes of evolution. After a period of evolution in each lab, all strains of each species were competed with one another. In yeast, the most successful strategies were those that used mating, underscoring the importance of sex in evolution. In bacteria, the fittest strain used a strategy based on exploration of different mutation rates. Different strategies displayed variable levels of performance and stability across additional challenges and conditions. This study therefore uncovers principles of effective experimental evolutionary regimens and might prove useful also for biotechnological developments of new strains and for understanding natural strategies in evolutionary arms races between species. Evolthon constitutes a model for community-based scientific exploration that encourages creativity and cooperation.
[All authors]
2018
-
(2018) Trends in Genetics. 34, 11, p. 832-845 Abstract
The ability of cells to grow and divide, differentiate and function, and even senesce is dependent on the fine-tuning of both gene and protein expression. Protein concentration in the cell is regulated not only at the transcriptional and post-translational levels, but also at the level of translation. Ribosomes, the molecular machines behind translation, were once considered to be an invariant driving force behind protein expression. However, studies over the past decade paint a rather different picture; namely, that ribosomes constitute an additional layer of regulatory control that might define which subsets of mRNAs are translated, to what extent, and to what purpose. Recent works summarized herein directly implicate ribosome heterogeneity and, in particular, ribosomal protein (RP) paralog specificity in regulating mRNA translation and control of the cellular translatome.
-
(2018) Journal of Cell Biology. 217, 1, p. 117-126 Abstract
Genome duplication in eukaryotes created paralog pairs of ribosomal proteins (RPs) that show high sequence similarity/identity. However, individual paralogs can confer vastly different effects upon cellular processes, e.g., specific yeast paralogs regulate actin organization, bud site selection, and mRNA localization, although how specificity is conferred is unknown. Changes in the RP composition of ribosomes might allow for specialized translation of different subsets of mRNAs, yet it is unclear whether specialized ribosomes exist and if paralog specificity controls translation. Using translatome analyses, we show that the translation of mitochondrial proteins is highly down-regulated in yeast lacking RP paralogs required for normal mitochondrial function (e.g., RPL1b). Although RPL1a and RPL1b encode identical proteins, Rpl1b-containing ribosomes confer more efficient translation of respiration-related proteins. Thus, ribosomes varying in RP composition may confer specialized functions, and RP paralog specificity defines a novel means of translational control.Errata: The first version of
this article published online included errors in Fig. 3 d that
occurred during production. The x axis of the two graphs in panel d had labels
for \u201crpl2aΔ\u201d and \u201crpl2bΔ\u201d that should have been \u201crpl1aΔ\u201d
and \u201crpl1bΔ,\u201d respectively. -
(2018) Bio-Protocol. 8, 21, 3070. Abstract
Transcription and RNA decay play critical roles in the process of gene expression and the ability to accurately measure cellular mRNA levels is essential for understanding this regulation. Here, we describe a single-molecule fluorescent in situ hybridization (smFISH) method (as performed in Haimovich et al., 2017) that detects single RNA molecules in individual cells. This technique employs multiple single-stranded, fluorescent labeled, short DNA probes that hybridize to target RNAs in fixed cells, allowing for both the quantification and localization of cytoplasmic and nuclear RNAs at the single-cell level and single-molecule resolution. Analyzing smFISH data provides absolute quantitative data of the number of cytoplasmic ("mature") mRNAs, the number of nascent RNA molecules at distinct transcription sites, and the spatial localization of these RNAs in the cytoplasm and/or nucleoplasm.
-
(2018) Molecular Biology of the Cell. 29, 8, p. 948-963 Abstract
Cdc48/p97 is known primarily for the retrotranslocation of misfolded proteins in endoplasmic reticulum (ER)-associated protein degradation (ERAD). Here we uncover a novel function for both Cdc48 and the conserved ubiquitin-associated/ubiquitin-like ubiquitin receptor (ubiquilin) proteins in yeast (e.g., Ddi1, Dsk2, and Rad23), which deliver ubiquitinated proteins to the proteasome for degradation. We show that Cdc48, its core adaptors Npl4 and Ufd1, and the ubiquilins confer the constitutive anterograde delivery of carboxypeptidase S (Cps1), a membranal hydrolase, to the multivesicular body (MVB) and vacuolar lumen. Cdc48 and Ddi1 act downstream of Rsp5-dependent Cps1 ubiquitination to facilitate the disassembly of insoluble Cps1 oligomers and upstream of ESCRT-0 to facilitate the entry of soluble protein into the MVB. Consequentially, detergent-insoluble Cps1 accumulates in cells bearing mutations in CDC48, DDI1, and all three ubiquilins (ddi1 Delta, dsk2 Delta, rad23 Delta). Thus, Cdc48 and the ubiquilins have ERAD- and proteasome-independent functions in the anterograde delivery of specific proteins to the yeast vacuole for proteolytic activation. As Cdc48/p97 and the ubiquilins are major linkage groups associated with the onset of human neurodegenerative disease (e.g., amytrophic lateral sclerosis, Alzheimer's, and Paget's disease of the bone), there may be a connection between their involvement in anterograde protein sorting and disease pathogenesis.
2017
-
(2017) Proceedings Of The National Academy Of Sciences Of The United States Of America-Physical Sciences. 114, 46, p. E9873-E9882 Abstract
RNAs have been shown to undergo transfer between mammalian cells, although the mechanism behind this phenomenon and its overall importance to cell physiology is not well understood. Numerous publications have suggested that RNAs (microRNAs and incomplete mRNAs) undergo transfer via extracellular vesicles (e.g., exosomes). However, in contrast to a diffusion-based transfer mechanism, we find that full-length mRNAs undergo direct cell-cell transfer via cytoplasmic extensions characteristic of membrane nanotubes (mNTs), which connect donor and acceptor cells. By employing a simple coculture experimental model and using single-molecule imaging, we provide quantitative data showing that mRNAs are transferred between cells in contact. Examples of mRNAs that undergo transfer include those encoding GFP, mouse beta-actin, and human Cyclin D1, BRCA1, MT2A, and HER2. We show that intercellular mRNA transfer occurs in all coculture models tested (e.g., between primary cells, immortalized cells, and in cocultures of immortalized human and murine cells). Rapid mRNA transfer is dependent upon actin but is independent of de novo protein synthesis and is modulated by stress conditions and gene-expression levels. Hence, this work supports the hypothesis that full-length mRNAs undergo transfer between cells through a refined structural connection. Importantly, unlike the transfer of miRNA or RNA fragments, this process of communication transfers genetic information that could potentially alter the acceptor cell proteome. This phenomenon may prove important for the proper development and functioning of tissues as well as for host-parasite or symbiotic interactions.
-
(2017) Mitochondria: Practical Protocols. 2nd Edition ed. p. 197-216 Abstract
Mitochondria are thought to have evolved from ancestral proteobacteria and, as a result of symbiosis, became an indispensable organelle in all eukaryotic cells. Mitochondria perform essential functions that provide the cell with ATP, amino acids, phospholipids, and both heme and iron-sulfur clusters. However, only 1% of mitochondrial proteins are encoded by the mitochondrial genome, while the remaining 99% are encoded in the nucleus. This raises a logistical challenge to the cell, as these nuclear-encoded proteins have to be translated, delivered to the mitochondrial surface, and translocated to its various compartments. Over the past decade, it was shown that subsets of mRNAs encoding mitochondrial proteins (mMPs) are localized to the mitochondrial surface in both yeast and mammalian cells. Moreover, factors (e.g., RNA-binding proteins) have been discovered that facilitate mMP targeting, and their loss leads to RNA mislocalization and defects in mitochondrial function (e.g., deficient respiration). Therefore, there is a demand in the field of mitochondrial biology to accurately measure mMP localization to the mitochondrial surface. In this chapter, we describe two techniques that allow for the visualization of mMPs using single-molecule fluorescent in situ hybridization and preparation of a highly enriched mitochondrial fraction followed by quantitative real-time PCR. Together, these techniques constitute powerful tools to link changes in mMP trafficking to defects in mitochondrial physiology.
2016
-
(2016) Molecular Biology of the Cell. 27, 25, p. 851-851 B1079. Abstract
The interplay between mRNAs and proteins plays a crucial role in the regulation of cell morphology and physiology, and recent studies reveal that many proteins can serve as RNAbinding proteins (RBPs) in addition to their previously characterized roles. By using the yeast as an organism model, we have tagged endogenously expressed mRNAs of interest with MS2 aptamer, followed by biochemical pulldown and mass spectrometry analysis to reveal the identity of known proteins that act as RBPs in the trafficking of specific mRNAs. Here, we present two examples of novel RBPs essential for cellular physiological functions in yeast.First, we have found that the COPI vesicle coat complex is necessary for the localization of mRNAs encoding mitochondrial proteins (mMPs) to mitochondria and for mitochondrial function. Inactivation of any one of the COPI proteins leads to reduced mMP binding to COPI itself, resulting in the dissociation ofmMPs from mitochondria, a reduction in mitochondrial membrane potential, a decrease in protein import in vivo and in vitro, and severe deficiencies in mitochondrial respiration. Using a model mMP (OXA1), we observed that COPI inactivation (or mutation of the potential COPIinteraction site) led to altered mRNA localization and impaired cellular respiration (Zabezhinsky et al., Cell Rep (2016) 15:540549).Second, we identified a histone 4 (Hhf1) as a protein that binds to STE2 mRNA, which encodes receptor for αfactor and is involved in cellcell mating. Deletion of either one of the two paralogs of histone 4 led to a twofold reduction in mating efficiency, while the deletion of two specific histone acetyl transferases also had the same effect. By applying the CRISPR/Cas9 system in yeast we successfully mutated the endogenous acetylation sites of HHF1 and saw a marked reduction in mating efficiency. Furthermore, mutation of the acetylation sites in Hhf1 showed reduced binding to STE2 mRNA. Together, our approach shows that endogenous mRNA tagging followed by the pulldown and analysis of bound protein can lead to surprising new insights into the functions of wellknown proteins.
-
(2016) RNA. 22, 5, p. 660-666 Abstract
The MS2 system has been extensively used to visualize single mRNA molecules in live cells and follow their localization and behavior. In their Letter to the Editor recently published, Garcia and Parker suggest that use of the MS2 system may yield erroneous mRNA localization results due to the accumulation of 3 decay products. Here we cite published works and provide new data which demonstrate that this is not a phenomenon general to endogenously expressed MS2-tagged transcripts, and that some of the results obtained in their study could have arisen from artifacts of gene expression.
-
(2016) Biochimica et Biophysica Acta - Molecular Cell Research. 1863, 5, p. 911-921 Abstract
Peroxisomes are distinct membrane-enclosed organelles involved in the β-oxidation of fatty acids and synthesis of ether phospholipids (e.g. plasmalogens), as well as cholesterol and its derivatives (e.g. bile acids). Peroxisomes comprise a distinct and highly segregated subset of cellular proteins, including those of the peroxisome membrane and the interior matrix, and while the mechanisms of protein import into peroxisomes have been extensively studied, they are not fully understood. Here we will examine the potential role of RNA trafficking and localized translation on protein import into peroxisomes and its role in peroxisome biogenesis and function. Given that RNAs encoding peroxisome biogenesis (PEX) and matrix proteins have been found in association with the endoplasmic reticulum and peroxisomes, it suggests that localized translation may play a significant role in the import pathways of these different peroxisomal constituents. This article is part of a Special Issue entitled: Peroxisomes edited by Ralf Erdmann.
-
(2016) Cell Reports. 15, 3, p. 540-549 Abstract
Nuclear-encoded mRNAs encoding mitochondrial proteins (mMPs) can localize directly to the mitochondrial surface, yet how mMPs target mitochondria and whether RNA targeting contributes to protein import into mitochondria and cellular metabolism are unknown. Here, we show that the COPI vesicle coat complex is necessary for mMP localization to mitochondria and mitochondrial function. COPI inactivation leads to reduced mMP binding to COPI itself, resulting in the dissociation of mMPs from mitochondria, a reduction in mitochondrial membrane potential, a decrease in protein import in vivo and in vitro, and severe deficiencies in mitochondrial respiration. Using a model mMP (OXA1), we observed that COPI inactivation (or mutation of the potential COPI-interaction site) led to altered mRNA localization and impaired cellular respiration. Overall, COPI-mediated mMP targeting is critical for mitochondrial protein import and function, and transcript delivery to the mitochondria or endoplasmic reticulum is regulated by cis-acting RNA sequences and trans-acting proteins.
2015
-
(2015) Cell Reports. 12, 11, p. 1876-1886 Abstract
Upon amino acid (AA) starvation and TOR inactivation, plasma-membrane-localized permeases rapidly undergo ubiquitination and internalization via the vacuolar protein sorting/multivesicular body (VPS-MVB) pathway and are degraded in the yeast vacuole. We now show that specific Golgi proteins are also directed to the vacuole under these conditions as part of a Golgi quality-control (GQC) process. The degradation of GQC substrates is dependent upon ubiquitination by the defective-for-SREBP-cleavage (DSC) complex, which was identified via genetic screening and includes the Tul1 E3 ligase. Using a model GQC substrate, GFP-tagged Yif1, we show that vacuolar targeting necessitates upregulation of the VPS pathway via proteasome-mediated degradation of the initial endosomal sorting complex required for transport, ESCRT-0, but not downstream ESCRT components. Thus, early cellular responses to starvation include the targeting of specific Golgi proteins for degradation, a phenomenon reminiscent of the inactivation of BTN1, the yeast Batten disease gene ortholog.
2013
-
(2013) Molecular Biology of the Cell. 24, 19, p. 3069-3084 Abstract
mRNAs encoding secreted/membrane proteins (mSMPs) are believed to reach the endoplasmic reticulum (ER) in a translation-dependent manner to confer protein translocation. Evidence exists, however, for translation- and signal recognition particle (SRP)-inde-pendent mRNA localization to the ER, suggesting that there are alternate paths for RNA delivery. We localized endogenously expressed mSMPs in yeast using an aptamer-based RNA-tagging procedure and fluorescence microscopy. Unlike mRNAs encoding polarity and secretion factors that colocalize with cortical ER at the bud tip, mSMPs and mRNAs encoding soluble, nonsecreted, nonpolarized proteins localized mainly to ER peripheral to the nucleus (nER). Synthetic nontranslatable uracil-rich mRNAs were also demonstrated to colocalize with nER in yeast. This mRNA-ER association was verified by subcellular fractionation and reverse transcription-PCR, single-molecule fluorescence in situ hybridization, and was not inhibited upon SRP inactivation. To better understand mSMP targeting, we examined aptamer-tagged USE1, which encodes a tail-anchored membrane protein, and SUC2, which encodes a soluble secreted enzyme. USE1 and SUC2 mRNA targeting was not abolished by the inhibition of translation or removal of elements involved in translational control. Overall we show that mSMP targeting to the ER is both translation- and SRP-independent, and regulated by cis elements contained within the message and trans-acting RNA-binding proteins (e.g., She2, Puf2).
2012
-
(2012) Cell Reports. 1, 5, p. 483-494 Abstract
mRNAs encoding polarity and secretion factors (POLs) target the incipient bud site in yeast for localized translation during division. In pheromone-treated cells we now find that these mRNAs are also localized to the yeast-mating projection (shmoo) tip. However, in contrast to the budding program, neither the She2 nor She3 proteins are involved. Instead, the Scp160 RNA-binding protein binds POL and mating pathway mRNAs and regulates their spatial distribution in a Myo4- and cortical ER-dependent fashion. RNA binding by Scp160 is stimulated by activation of Gpa1, the G protein α subunit regulated by the pheromone receptor, and is required for pheromone gradient sensing, as well as subsequent chemotropic growth and cell-cell mating. These effects are incurred independently of obvious changes in translation; thus, mRNA trafficking is required for chemotropism and completion of the mating program. This is, to our knowledge, the first demonstration of ligand-activated RNA targeting in the development of a simple eukaryote.
2011
-
(2011) Rna-A Publication Of The Rna Society. 17, 12, p. 2249-2255 Abstract
Protein localization within cells can be achieved by the targeting and localized translation of mRNA. Yet, our understanding of the dynamics of mRNA targeting and protein localization, and of how general this phenomenon is, is not clear. Plasmid-based expression systems have been used to visualize exogenously expressed mRNAs and proteins; however, these methods typically produce them at levels greater than endogenous and can result in mislocalization. Hence, a method that allows for the simultaneous visualization of endogenous mRNAs and their translation products in living cells is needed. We previously developed a method (m-TAG) to localize endogenously expressed mRNAs in yeast by chromosomal insertion of the MS2 aptamer sequence between the open-reading frame (ORF) and 3 UTR of any gene. Upon coexpression with the MS2 RNAbinding coat protein (MS2-CP) fused with GFP, the aptamer-tagged mRNAs bearing their 3 UTRs are localized using fluorescence microscopy. Here we describe an advanced method (mp-TAG) that allows for the simultaneous visualization of both endogenously expressed mRNAs and their translation products in living yeast for the first time. Homologous recombination is used to insert the mCherry gene and MS2-CP binding sites downstream from any ORF, in order to localize protein and mRNA, respectively. As proof of the concept, we tagged ATP2 as a representative gene and demonstrated that endogenous ATP2 mRNA and protein localize to mitochondria, as shown previously. In addition, we demonstrate that tagged proteins like Hhf2, Vph1, and Yef3 localize to their expected subcellular location, while the localization of their mRNAs is revealed for the first time. Published by Cold Spring Harbor Laboratory Press.
-
(2011) Journal of Cell Biology. 195, 2, p. 203-215 Abstract
The human Batten disease gene CLN3 and yeast orthologue BTN1 encode proteins of unclear function. We show that the loss of BTN1 phenocopies that of BTN2, which encodes a retromer accessory protein involved in the retrieval of specific cargo from late endosomes (LEs) to the Golgi. However, Btn1 localizes to Golgi and regulates soluble N-ethyl-maleimide sensitive fusion protein attachment protein receptor (SNARE) function to control retrograde transport. Specifically, BTN1 overexpression and deletion have opposing effects on phosphorylation of the Sed5 target membrane SNARE, on Golgi SNARE assembly, and on Golgi integrity. Although Btn1 does not interact physically with SNAREs, it regulates Sed5 phosphorylation by modulating Yck3, a palmitoylated endosomal kinase. This may involve modification of the Yck3 lipid anchor, as substitution with a transmembrane domain suppresses the deletion of BTN1 and restores trafficking. Correspondingly, deletion of YCK3 mimics that of BTN1 or BTN2 with respect to LE-Golgi retrieval. Thus, Btn1 controls retrograde sorting by regulating SNARE phosphorylation and assembly, a process that may be adversely affected in Batten Disease patients.
-
(2011) Rna-A Publication Of The Rna Society. 17, 8, p. 1551-1565 Abstract
Targeted mRNA localization is a likely determinant of localized protein synthesis. To investigate whether mRNAs encoding mitochondrial proteins (mMPs) localize to mitochondria and, thus, might confer localized protein synthesis and import, we visualized endogenously expressed mMPs in vivo for the first time. We determined the localization of 24 yeast mMPs encoding proteins of the mitochondrial matrix, outer and inner membrane, and intermembrane space and found that many mMPs colocalize with mitochondria in vivo. This supports earlier cell fractionation and microarray-based studies that proposed mMP association with the mitochondrial fraction. Interestingly, a number of mMPs showed a dependency on the mitochondrial Puf3 RNA-binding protein, as well as nonessential proteins of the translocase of the outer membrane (TOM) complex import machinery, for normal colocalization with mitochondria. We examined the specific determinants of ATP2 and OXA1 mRNA localization and found a mutual dependency on the 3 UTR, Puf3, Tom7, and Tom70, but not Tom20, for localization. Tom6 may facilitate the localization of specific mRNAs as OXA1, but not ATP2, mRNA was mislocalized in tom6D cells. Interestingly, a substantial fraction of OXA1 and ATP2 RNA granules colocalized with the endoplasmic reticulum (ER) and a deletion in MDM10, which mediates mitochondria-ER tethering, resulted in a significant loss of OXA1 mRNA localization with ER. Finally, neither ATP2 nor OXA1 mRNA targeting was affected by a block in translation initiation, indicating that translation may not be essential for mRNA anchoring. Thus, endogenously expressed mRNAs are targeted to the mitochondria in vivo, and multiple factors contribute to mMP localization. Published by Cold Spring Harbor Laboratory Press.
-
(2011) Molecular Biology of the Cell. 22, 10, p. 1648-1663 Abstract
Yeast Btn2 facilitates the retrieval of specific proteins from late endosomes (LEs) to the Golgi, a process that may be adversely affected in Batten disease patients. We isolated the putative yeast orthologue of a human complex I deficiency gene, designated here as BTN3, as encoding a Btn2-interacting protein and negative regulator. First, yeast overexpressing BTN3 phenocopy the deletion of BTN2 and mislocalize certain trans-Golgi proteins, like Kex2 and Yif1, to the LE and vacuole, respectively. In contrast, the deletion of BTN3 results in a tighter pattern of protein localization to the Golgi. Second, BTN3 overexpression alters Btn2 localization from the IPOD compartment, which correlates with a sharp reduction in Btn2-mediated [URE3] prion curing. Third, Btn3 and the Snc1 v-SNARE compete for the same binding domain on Btn2, and this competition controls Btn2 localization and function. The inhibitory effects upon protein retrieval and prion curing suggest that Btn3 sequesters Btn2 away from its substrates, thus down-regulating protein trafficking and aggregation. Therefore Btn3 is a novel negative regulator of intracellular protein sorting, which may be of importance in the onset of complex I deficiency and Batten disease in humans.
-
(2011) Rna Detection And Visualization. p. 237-247 Abstract
Localized mRNA translation is involved in cell-fate determination, polarization, and morphogenesis in eukaryotes. While various tools are available to examine mRNA localization, no easy and quick method has allowed for the visualization of endogenously expressed mRNAs in vivo. We describe a simple method (m-TAG) for PCR-based chromosomal gene tagging that uses homologous recombination to insert binding sites for the RNA-binding MS2 coat protein (MS2-CP) between the coding region and 3-untranslated region of any yeast gene. Upon co-expression of MS2-CP fused with GFP, specific endogenously expressed mRNAs can be visualized in vivo for the first time. This method allows for the easy examination of mRNA localization using fluorescence microscopy and leaves the yeast cells amenable for further genetic analysis.
-
(2011) Rna Detection And Visualization. Gerst J.(eds.). p. 323-333 Abstract
Targeted mRNA localization to distinct subcellular sites occurs throughout the eukaryotes and presumably allows for the localized translation of proteins near their site of function. Specific mRNAs have been localized in cells using a variety of reliable methods, such as fluorescence in situ hybridization with labeled RNA probes, mRNA tagging using RNA aptamers and fluorescent proteins that recognize these aptamers, and quenched fluorescent RNA probes that become activated upon binding to mRNAs. However, fluorescence-based RNA localization studies can be strengthened when coupled with cell fractionation and membrane isolation techniques in order to identify mRNAs associated with specific organelles or other subcellular structures. Here we describe a novel method to isolate mRNAs associated with peroxisomes in the yeast, Saccharomyces cerevisiae. This method employs a combination of density gradient centrifugation and affinity purification to yield a highly enriched peroxisome fraction suitable for RNA isolation and reverse transcription-polymerase chain reaction detection of mRNAs bound to peroxisome membranes. The method is presented for the analysis of peroxisome-associated mRNAs; however it is applicable to studies on other subcellular compartments.
-
(2011) Rna Detection And Visualization. Gerst J.(eds.). p. 387-406 Abstract
RNA metabolism involves regulatory processes, such as transcription, splicing, nuclear export, transport and localization, association with sites of RNA modification, silencing and decay, and necessitates a wide variety of diverse RNA-interacting proteins. These interactions can be direct via RNA-binding proteins (RBPs) or indirect via other proteins and RNAs that form ribonucleoprotein complexes that together control RNA fate. While pull-down methods for the isolation of known RBPs are commonly used, strategies have also been described for the direct isolation of messenger RNAs (mRNAs) and their associated factors. The latter techniques allow for the identification of interacting proteins and RNAs, but most suffer from problems of low sensitivity and high background. Here we describe a simple and highly effective method for RNA purification and identification (RaPID) that allows for the isolation of specific mRNAs of interest from yeast and mammalian cells, and subsequent analysis of the associated proteins and RNAs using mass spectrometry and reverse transcription-PCR, respectively. This method employs the MS2 coat RBP fused to both GFP and streptavidin-binding protein to precipitate MS2 aptamer-tagged RNAs using immobilized streptavidin.
2010
-
(2010) Rna-A Publication Of The Rna Society. 16, 11, p. 2277-2290 Abstract
Intracellular mRNA targeting and localized translation are potential determinants for protein localization. To facilitate targeting, mRNAs possess specific cis-acting sequence motifs that are recognized by trans-acting RNA-binding proteins (RBPs). While many mRNAs are trafficked, our knowledge of the RBPs involved and presence of additional transcripts within these ribonucleoprotein (RNP) complexes is limited. To facilitate the identification of RBPs and transcripts that bind to specific mRNAs, we developed RNA-binding protein purification and identification (RaPID), a novel technique that allows for the affinity purification of MS2 aptamer-tagged mRNAs and subsequent detection of bound RBPs and transcripts using mass-spectometry and RT-PCR, respectively. RaPID effectively isolated specific mRNAs from both yeast and mammalian cells, and identified known mRNA-RBP interactions (e.g., ASH1-She2; β-Actin-IMP1). By isolating tagged OXA1 mRNA using RaPID, we could identify a yeast COPI subunit (i.e., Sec27) as a candidate interacting protein. This finding was strengthened by the observation that a portion of OXA1 mRNA was delocalized in a sec27-1 temperature-sensitive mutant at restrictive temperatures. Finally, RaPID could also be used to show biochemically the coexistence of different RNA species within the same RNP complex (e.g., coprecipitation of the yeast SRO7, WSC2, SEC3, and IST2 mRNAs with ASH1 mRNA) for the first time.
-
(2010) Trends in Biochemical Sciences. 35, 8, p. 459-469 Abstract
Translation-coupled protein translocation requires that mRNAs encoding secreted and membrane proteins (mSMPs) reach the ER membrane. The classical view is that the signal recognition particle (SRP) pathway delivers translating signal sequence-containing proteins to the SRP receptor present on the ER surface and engages the translocation machinery. However, recent studies demonstrate both SRP- and translation-independent mRNA recruitment to the ER, and that mRNAs encoding non-signal sequence-containing cytosolic proteins (mCPs) might be full-time residents of ER membranes. Furthermore, translation-independent cis-acting sequence elements present in both mCPs and mSMPs appear to govern the ability of mRNAs to associate with ER. Thus, a more complex picture of how and why mRNAs target the ER is emerging.
2009
-
(2009) Trends in Cell Biology. 19, 12, p. 677-684 Abstract
Cell polarity is necessary for cell division, morphogenesis and motility in eukaryotes, and is determined by dynamic control of the cytoskeleton and secretory pathway to promote directional growth. In yeast, three essential and tightly-regulated processes orchestrate polarization and facilitate bud growth. These processes include phosphoinositide (PI) signaling, Rho GTPase regulation of the actin cytoskeleton, and exocytosis. As yet, the interplay between these different processes is unclear, and two main models (Spatial Landmark and Allosteric Local Activation) have been proposed for Rho GTPase control of polarization in yeast. Here, we summarize the inter-relationship between these growth processes and present a more unified model, the Exocytic Signal model, which proposes that exocytosis and actin regulation are fully integrated events mediated by PI signaling.
-
(2009) Proceedings of the National Academy of Sciences of the United States of America. 106, 47, p. 19848-19853 Abstract
Targeted mRNA trafficking and local translation may play a significant role in controlling protein localization. Here we examined for the first time the localization of all (≈50) mRNAs encoding peroxisomal proteins (mPPs) involved in peroxisome biogenesis and function. By using the bacteriophage MS2-CP RNA-binding protein (RBP) fused to multiple copies of GFP, we demonstrated that >40 endogenously expressed mPPs tagged with the MS2 aptamer form fluorescent RNA granules in vivo. The use of different RFP-tagged organellar markers revealed 3 basic patterns of mPP granule localization: to peroxisomes, to the endoplasmic reticulum (ER), and nonperoxisomal. Twelve mPPs (i.e., PEX1, PEX5, PEX8, PEX11-15, DCI1, NPY1, PCS60, and POX1) had a high percentage (52%-80%) of mRNA colocalization with peroxisomes. Thirteen mPPs (i.e., AAT2, PEX6, MDH3, PEX28, etc.) showed a low percentage (30%-42%) of colocalization, and 1 mPP (PEX3) preferentially localized to the ER. The mPPs of the nonperoxisomal pattern (i.e., GPD1, PCD1, PEX7) showed ≪30% colocalization. mPP association with the peroxisome or ER was verified using cell fractionation and RT-PCR analysis. A model mPP, PEX14 mRNA, was found to be in close association with peroxisomes throughout the cell cycle, with its localization depending in part on the 3-UTR, initiation of translation, and the Puf5 RBP. The different patterns of mPP localization observed suggest that multiple mechanisms involved in mRNA localization and translation may play roles in the importation of protein into peroxisomes.
-
(2009) Molecular Biology of the Cell. 20, 15, p. 3583-3597 Abstract
The actin cytoskeleton rapidly depolarizes in yeast secretory (sec) mutants at restrictive temperatures. Thus, an unknown signal conferred upon secretion is necessary for actin polarity and exocytosis. Here, we show that a phosphatidylinositol (PI) transfer protein, Sfh5, and a phosphatidylinositol-4- phosphate 5-kinase, Mss4, facilitate Cdc42 activation to concomitantly regulate both actin and protein trafficking. Defects in Mss4 function led to actin depolarization, an inhibition of secretion, reduced levels of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] in membranes, mislocalization of a pleckstrin homology domain fused to green fluorescent protein, and the mislocalization of Cdc42. Similar defects were observed in sec, myo2-66, and cdc42-6 mutants at elevated temperatures and were rescued by the overexpression of MSS4. Likewise, the overexpression of SFH5 or CDC42 could ameliorate these defects in many sec mutants, most notably in sec3Δ cells, indicating that Cdc42-mediated effects upon actin and secretion do not necessitate Sec3 function. Moreover, mutation of the residues involved in PI binding in Sfh5 led to the mislocalization and loss of function of both Sfh5 and Cdc42. Based upon these findings, we propose that the exocytic signal involves PI delivery to the PI kinases (i.e., Mss4) by Sfh5, generation of PI(4,5)P 2, and PI(4,5)P2-dependent regulation of Cdc42 and the actin cytoskeleton.
-
(2009) Nature protocols. 4, 9, p. 1274-1284 Abstract
This protocol describes m-TAG, a novel method for the visualization of endogenously expressed mRNAs in live yeast (Saccharomyces cerevisiae). First, a gene of interest is tagged with multiple binding sites for the RNA-binding MS2 coat protein (MS2-CP), using a PCR-based genomic-tagging strategy and homologous recombination. Next, MS2-CP fused to GFP(x3) is expressed in cells; binding of this fusion protein to the tagged mRNA yields an RNA granule that can be visualized by fluorescence microscopy. While existing methods necessitate cell fixation (for in situ hybridization) or the detection of exogenously expressed mRNAs (from plasmids), or give transient signals (i.e., with fluorescent hybridization probes), m-TAG allows for the robust and stable visualization of endogenously expressed mRNAs in vivo and facilitates the study of mRNA dynamics under different growth conditions. The m-TAG procedure is simple, easy to perform and takes
2008
-
(2008) Molecular Biology of the Cell. 19, 9, p. 3625-3637 Abstract
Ddi1/Vsm1 is an ubiquitin receptor involved in regulation of the cell cycle and late secretory pathway in Saccharomyces cerevisiae. Ddi1 possesses three domains: an NH2-terminal ubiquitin-like domain (UBL), a COOH-terminal ubiquitin-associated domain (UBA), and a retroviral aspartyl-protease domain (RVP). Here, we demonstrate the domains involved in homodimerization, checkpoint regulation, localization, and t-SNARE binding. The RVP domain is required for protein homodimerization, whereas the UBL and UBA domains are required for rescue of the pds1-128 checkpoint mutant and enrichment of GFP-Ddi1 in the nucleus. A mutation in aspartate-220, which is necessary for putative aspartyl-protease function, abolished the rescue of pds1-128 cells but not homodimerization. Thus, Ddi1 catalytic activity may be required for checkpoint regulation. The Sso1 t-SNARE-interacting domain maps to residues 344-395 and undergoes phosphorylation on threonines T346 and T348. T348 is necessary for Sso binding, and phosphorylation is important for function, because mutations that lessen phosphorylation (e.g., Ddi1T346A, Ddi1T348A) are unable to facilitate growth of the sec9-4 t-SNARE mutant. In contrast, the overproduction of phosphorylatable forms of Ddi1 (e.g., Ddi1, Ddi 1S341A) rescue the growth of sec9-4 cells similar to Sso1 overproduction. Thus, Ddi1 participates in multiple cellular processes via its different domains and phosphorylation may regulate exocytic functions.
-
(2008) Trends in Cell Biology. 18, 2, p. 68-76 Abstract
In eukaryotes, mRNAs encoding secreted and integral membrane proteins are targeted to the endoplasmic reticulum (ER) to facilitate translation and protein translocation into the ER lumen. However, mRNAs encoding cytosolic proteins also associate with ER membranes in yeast, plants and animal cells. mRNAs encoding both cytosolic and secreted proteins have been observed in association with the cortical ER (cER) network, which consists of interconnected tubular and sheet-like structures that extend to the plasma membrane and to sites of polarized growth. This physical association enables cytoskeleton-mediated co-trafficking and anchoring of cER-mRNA, which might regulate protein synthesis in areas of new growth (i.e. during cell division in yeast), or enable confined spatial responses to environmental stimuli (i.e. during synaptic remodeling or in cases of neuronal injury).
-
2007
-
(2007) Nature Methods. 4, 5, p. 409-412 Abstract
mRNA localization may be an important determinant for protein localization. We describe a simple PCR-based genomic-tagging strategy (m-TAG) that uses homologous recombination to insert binding sites for the RNA-binding MS2 coat protein (MS2-CP) between the coding region and 3 untranslated region (UTR) of any yeast gene. Upon coexpression of MS2-CP fused with GFP, we demonstrate the localization of endogenous mRNAs (ASH1, SRO7, PEX3 and OXA1) in living yeast (Saccharomyces cerevisiae).
-
(2007) Molecular and Cellular Biology. 27, 9, p. 3441-3455 Abstract
Polarized growth in the budding yeast Saccharomyces cerevisiae depends upon the asymmetric localization and enrichment of polarity and secretion factors at the membrane prior to budding. We examined how these factors (i.e., Cdc42, Sec4, and Sro7) reach the bud site and found that their respective mRNAs localize to the tip of the incipient bud prior to nuclear division. Asymmetric mRNA localization depends upon factors that facilitate ASH1 mRNA localization (e.g., the 3 untranslated region, She proteins 1 to 5, Puf6, actin cytoskeleton, and a physical association with She2). mRNA placement precedes protein enrichment and subsequent bud emergence, implying that mRNA localization contributes to polarization. Correspondingly, mRNAs encoding proteins which are not asymmetrically distributed (i.e., Snc1, Mso1, Tub1, Pex3, and Oxa1) are not polarized. Finally, mutations which affect cortical endoplasmic reticulum (ER) entry and anchoring in the bud (myo4Δ, sec3Δ, and srp101) also affect asymmetric mRNA localization. Bud-localized mRNAs, including ASH1, were found to cofractionate with ER microsomes in a She2- and Sec3-dependent manner; thus, asymmetric mRNA transport and cortical ER inheritance are connected processes in yeast.
-
(2007) Molecular and Cellular Biology. 27, 2, p. 605-621 Abstract
BTN2 gene expression in the yeast Saccharomyces cerevisiae is up-regulated in response to the deletion of BTN1, which encodes the ortholog of a human Batten disease protein. We isolated Btn2 as a Snc1 v-SNARE binding protein using the two-hybrid assay and examined its role in intracellular protein trafficking. We show that Btn2 is an ortholog of the Drosophila and mammalian Hook1 proteins that interact with SNAREs, cargo proteins, and coat components involved in endosome-Golgi protein sorting. By immunoprecipitation, it was found that Btn2 bound the yeast endocytic SNARE complex (e.g., Snc1 and Snc2 [Snc1/2], Tlg1, Tlg2, and Vti1), the Snx4 sorting nexin, and retromer (e.g., Vps26 and Vps35). In in vitro binding assays, recombinant His6-tagged Btn2 bound glutathione S-transferase (GST)-Snc1 and GST-Vps26. Btn2-green fluorescent protein and Btn2-red fluorescent protein colocalize with Tlg2, Snx4, and Vps27 to a compartment adjacent to the vacuole that corresponds to a late endosome. The deletion of BTN2 blocks Yif1 retrieval back to the Golgi apparatus, while the localization of Ste2, Fur4, Snc1, Vps10, carboxypeptidases Y (CPY) and S (CPS), Sed5, and Sec7 is unaltered in btn2Δ cells. Yif1 delivery to the vacuole was observed in other late endosome-Golgi trafficking mutants, including ypt6Δ, snx4Δ, and vps26Δ cells. Thus, Btn2 facilitates specific protein retrieval from a late endosome to the Golgi apparatus, a process which may be adversely affected in patients with Batten disease.
-
(2007) Molecular and Cellular Biology. 27, 2, p. 526-540 Abstract
Although COPI function on the early secretory pathway in eukaryotes is well established, earlier studies also proposed a nonconventional role for this coat complex in endocytosis in mammalian cells. Here we present results that suggest an involvement for specific COPI subunits in the late steps of endosomal protein sorting in Saccharomyces cerevisiae. First, we found that carboxypeptidase Y (CPY) was partially missorted to the cell surface in certain mutants of the COPIB subcomplex (COPIb; Sec27, Sec28, and possibly Sec33), which indicates an impairment in endosomal transport. Second, integral membrane proteins destined for the vacuolar lumen (i.e., carboxypeptidase S [CPS1]; Fur4, Ste2, and Ste3) accumulated at an aberrant late endosomal compartment in these mutants. The observed phenotypes for COPIb mutants resemble those of class E vacuolar protein sorting (vps) mutants that are impaired in multivesicular body (MVB) protein sorting and biogenesis. Third, we observed pliysical interactions and colocalization between COPIb subunits and an MVB-associated protein, Vps27. Together, our findings suggest that certain COPI subunits could have a direct role in vacuolar protein sorting to the MVB compartment.
2006
-
(2006) Molecular Biology of the Cell. 17, 4, p. 1845-1858 Abstract
Gcs1 is an Arf GTPase-activating protein (Arf-GAP) that mediates Golgi-ER and post-Golgi vesicle transport in yeast. Here we show that the Snc1,2 v-SNAREs, which mediate endocytosis and exocytosis, interact physically and genetically with Gcs1. Moreover, Gcs1 and the Snc v-SNAREs colocalize to subcellular structures that correspond to the trans-Golgi and endosomal compartments. Studies performed in vitro demonstrate that the Snc-Gcs1 interaction results in the efficient binding of recombinant Arf1Δ17N-Q71L to the v-SNARE and the recruitment of purified coatomer. In contrast, the presence of Snc had no effect on Gcs1 Arf-GAP activity in vitro, suggesting that v-SNARE binding does not attenuate Arf1 function. Disruption of both the SNC and GCS1 genes results in synthetic lethality, whereas overexpression of either SNC gene inhibits the growth of a distinct subset of COPI mutants. We show that GFP-Snc1 recycling to the trans-Golgi is impaired in gcs1Δ cells and these COPI mutants. Together, these results suggest that Gcs1 facilitates the incorporation of the Snc v-SNAREs into COPI recycling vesicles and subsequent endosome-Golgi sorting in yeast.
[All authors]
2005
-
(2005) Journal of Biological Chemistry. 280, 40, p. 34033-34041 Abstract
The yeast exocytic SNARE complex consists of one molecule each of the Sso1/2 target SNAREs, Snc1/2 vesicular SNAREs, and the Sec9 target SNARE, which form a fusion complex that is conserved in evolution. Another protein, Sec1, binds to the SNARE complex to facilitate assembly. We show that Mso1, a Sec1-interacting protein, also binds to the SNARE complex and plays a role in mediating Sec1 functions. Like Sec1, Mso1 bound to SNAREs in cells containing SNARE complexes (i.e. wild-type, sec1-1, and sec18-1 cells), but not in cells in which complex formation is inhibited (i.e. sec4-8 cells). Nevertheless, Mso1 remained associated with Seel even in sec4-8 cells, indicating that they act as a pair. Mso1 localized primarily to the plasma membrane of the bud when SNARE complex formation was not impaired but was mostly in the cytoplasm when assembly was prevented. Genetic studies suggest that Mso1 enhances Sec1 function while attenuating Sec4 GTPase function. This dual action may impart temporal regulation between Sec4 turnoff and Sec1-mediated SNARE assembly. Notably, a small region at the C terminus of Mso1 is conserved in the mammalian Munc13/Mint proteins and is necessary for proper membrane localization. Overexpression of Mso1 lacking this domain (Mso1-(1-193)) inhibited the growth of cells bearing an attenuated Sec4 GTPase. These results suggest that Mso1 is a component of the exocytic SNARE complex and a possible ortholog of the Munc13/Mint proteins.
-
(2005) Molecular Biology of the Cell. 16, 10, p. 4918-4930 Abstract
Previously, we demonstrated that the phosphorylation of t-SNAREs by protein kinase A (PKA) affects their ability to participate in SNARE complexes and to confer endocytosis and exocytosis in yeast. Here, we show that the presumed phosphorylation of a conserved membrane-proximal PKA consensus site (serine-317) in the Sed5 t-SNARE regulates endoplasmic reticulum (ER)-Golgi transport, as well as Golgi morphology. Sed5 is a phosphoprotein, and both alanine and aspartate substitutions in serine-317 directly affect intracellular protein trafficking. The aspartate substitution results in elaboration of the ER, defects in Golgi-ER retrograde transport, an accumulation of small transport vesicles, and the inhibition of growth of most cell types. In contrast, the alanine substitution has no deleterious effects upon transport and growth, but results in ordering of the Golgi into a structure reminiscent of mammalian apparatus. This structure seems to require the recycling of Sed5, because it was found not to occur in sec21-2 cells that are defective in retrograde transport. Thus, a cycle of Sed5 phosphorylation and dephosphorylation is required for normal t-SNARE function and may choreograph Golgi ordering and dispersal.
2004
-
(2004) Journal of Biological Chemistry. 279, 35, p. 36962-36971 Abstract
Both the delivery of secretory vesicles and asymmetric distribution of mRNA to the bud are dependent upon the actin cytoskeleton in yeast. Here we examined whether components of the exocytic apparatus play a role in mRNA transport. By screening secretion mutants in situ and in vivo, we found that all had an altered pattern of ASH1 mRNA localization. These included alleles of CDC42 and RHO3 (cdc42-6 and rho3-V51) thought to regulate specifically the fusion of secretory vesicles but were found to affect strongly the cytoskeleton as well. Most interestingly, mutations in late secretion-related genes not directly involved in actin regulation also showed substantial alterations in ASH1 mRNA distribution. These included mutations in genes encoding components of the exocyst (SEC10 and SEC15), SNARE regulatory proteins (SEC1, SEC4, and SRO7), SNAREs (SEC9 and SSO1/2), and proteins involved in Golgi export (PIK1 and YPT31/32). Importantly, prominent defects in the actin cytoskeleton were observed in all of these strains, thus implicating a known causal relationship between the deregulation of actin and the inhibition of mRNA transport. Our novel observations suggest that vesicular transport regulates the actin cytoskeleton in yeast (and not just vice versa) leading to subsequent defects in mRNA transport and localization.
-
2003
-
(2003) Molecular Biology of the Cell. 14, 8, p. 3114-3125 Abstract
We have shown that protein kinase A phosphorylation of t-SNAREs inhibits SNARE assembly and suppresses endo- and exocytosis in yeast. Herein, we show that protein kinase A phosphorylation of the Sso exocytic t-SNAREs promotes the binding of Vsm1, a potential SNARE regulator identified previously in our laboratory. Phosphorylation of Sso increases its affinity for Vsm1 by more than fivefold in vitro and both phosphorylated Sso1, as well as Sso1 bearing an aspartate substitution at position 79, interact tightly with Vsm1. Vsm1 binding is dependent upon the NH2-terminal autoinhibitory domain of Sso, and constitutively "open" forms of the t-SNARE show a reduction in Vsm1 binding in vivo. The substitution of serine-79 in Sso1 with an alanine residue or the treatment of yeast with C2-ceramide, which results in the dephosphorylation of serine-79, both inhibit Vsm1 binding in vivo. Importantly, Vsm1 binding to Sso seems to preclude Sso binding to its partner t-SNARE, Sec9, and vice versa. This is consistent with the idea that Vsm1 is an inhibitor of SNARE assembly in yeast. Thus, one way by which phosphorylation inhibits SNARE assembly could be by regulating the association of inhibitory factors that control the ability of t-SNAREs to form complexes in vivo.
-
(2003) Biochimica et Biophysica Acta - Molecular Cell Research. 1641, 2-3, p. 99-110 Abstract
SNAREs (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptors) are membrane-associated proteins that participate in the fusion of internal membranes in eukaryotic cells. SNAREs comprise three distinct and well-conserved families of molecules that act directly as membrane fusogens or, at the least, as elements that bring membranes into close apposition and allow for subsequent fusion events to occur. While the molecular events leading to fusion are still under debate, it is clear that a number of additional factors are required to bring about SNARE-mediated membrane fusion in vivo. Many of these factors, which collectively can be called SNARE regulators (e.g. Sec1/Munc18, synaptotagmin, GATE-16, LMA1, Munc13/UNC-13, synaptophysin, tomosyn, Vsm1, etc.), bind directly to SNAREs and are involved in the regulation of SNARE assembly as well as the ability of SNAREs to participate in trafficking events. In addition, recent studies have suggested a role for posttranslational modification (e.g., phosphorylation) in the regulation of SNARE functions. In this review the possible role of SNARE regulators in SNARE assembly and the involvement of SNARE phosphorylation in the regulation of intracellular membrane trafficking will be discussed.
2002
-
(2002) Journal of Biological Chemistry. 277, 38, p. 35274-35281 Abstract
The OS-9 gene maps to a region (q13-15) of chromosome 12 that is highly amplified in human osteosarcomas and encodes a protein of unknown function. Here we have characterized a homolog designated as YOS9 (YDR057w) from Saccharomyces cerevisiae. The yeast protein (Yos9) is a membrane-associated glycoprotein that localizes to the endoplasmic reticulum (ER). YOS9 interacts genetically with genes involved in ER-Golgi transport, particularly SEC34, whose temperature-sensitive mutant is rescued by YOS9 overexpression. Interestingly, Yos9 appears to play a direct role in the transport of glycosylphosphatidylinositol (GPI)-anchored proteins to the Golgi apparatus. Yos9 binds directly to Gas1 and Mkc7 and accelerates Gas1 transport and processing in cells overexpressing YOS9. Correspondingly, Gas1 processing is slowed in cells bearing a deletion in YOS9. No effect upon the transport and processing of non-GPI-anchored proteins (e.g. invertase and carboxypeptidase Y) was detected in cells either lacking or overexpressing Yos9. As Yos9 is not a component of the Emp24 complex, it may act as a novel escort factor for GPI-anchored proteins in ER-Golgi transport in yeast and possibly in mammals.
-
(2002) EMBO Journal. 21, 4, p. 602-614 Abstract
Yeast produce two classes of secretory vesicles (SVs) that differ in both density and cargo protein content. In late-acting secretory mutants (e.g. snc1ala43 and sec6-4), both low- (LDSV) and high-density (HDSV) classes of vesicles accumulate at restrictive temperatures. Here, we have found that disruptions in the genes encoding a dynamin-related protein (VPS1) or clathrin heavy chain (CHC1) abolish HDSV production, yielding LDSVs that contain all secreted cargos. Interestingly, disruption of the PEP12 gene, which encodes the t-SNARE that mediates all Golgi to prevacuolar compartment (PVC) transport, also abolishes HDSV production. In contrast, deletions in genes that selectively confer vacuolar hydrolase sorting to the PVC or protein transport to the vacuole (i.e. VPS34 and VAM3, respectively) have no effect. Thus, one branch of the secretory pathway in yeast involves an intermediate sorting compartment and has a specific requirement for clathrin and a dynamin-related protein in SV biogenesis.
-
(2002) Molecular Biology of the Cell. 13, 5, p. 1594-1607 Abstract
Earlier we demonstrated that activation of a ceramide-activated protein phosphatase (CAPP) conferred normal growth and secretion to yeast lacking their complement of exocytic v-SNAREs (Snc1,2) or bearing a temperature-sensitive mutation in an exocytic t-SNARE (Sso2). CAPP activation led to Sso dephosphorylation and enhanced the assembly of t-SNAREs into functional complexes. Thus, exocytosis in yeast is modulated by t-SNARE phosphorylation. Here, we show that endocytic defects in cells lacking the v- and t-SNAREs involved in endocytosis are also rescued by CAPP activation. Yeast lacking the Tlg1 or Tlg2 t-SNAREs, the Snc v-SNAREs, or both, undergo endocytosis after phosphatase activation. CAPP activation correlated with restored uptake of FM4-64 to the vacuole, the uptake and degradation of the Ste2 receptor after mating factor treatment, and the dephosphorylation and assembly of Tlg1,2 into SNARE complexes. Activation of the phosphatase by treatment with C2-ceramide, VBM/ELO gene inactivation, or by the overexpression of SIT4 was sufficient to confer rescue. Finally, we found that mutation of single PKA sites in Tlg1 (Ser31 to Ala31) or Tlg2 (Ser90 to Ala90) was sufficient to restore endocytosis, but not exocytosis, to snc cells. These results suggest that endocytosis is also modulated by t-SNARE phosphorylation in vivo.
2001
-
(2001) EMBO Journal. 20, 3, p. 411-421 Abstract
The role of protein phosphorylation in secretion is not well understood. Here we show that yeast lacking the Sncl,2 v-SNAREs, or bearing a temperature-sensitive mutation in the Sso2 t-SNARE, are rescued at restrictive conditions by the addition of ceramide precursors and analogs to the growth medium. Rescue results from dephosphorylation of the Sso t-SNAREs by a ceramide-activated type 2A protein phosphatase (Sit4) involved in cell cycle control. Sso t-SNARE dephosphorylation correlated with its assembly into complexes with the Sec9 t-SNARE, both in vitro and in vivo, and with an increase in protein trafficking and secretion in cells. SNARE complexes isolated under these conditions contained only Sso and Sec9, suggesting that a t-t-SNARE fusion complex is sufficient to confer exocytosis. Mutation of a single PKA site (Ser79 to Ala79) in Sso1 resulted in a decrease in phosphorylation and was sufficient to confer growth to snc cells at restrictive conditions. Thus, modulation of t-SNARE phosphorylation regulates SNARE complex assembly and membrane fusion in vivo.
1999
-
(1999) Cellular and Molecular Life Sciences. 55, 5, p. 707-734 Abstract
Eukaryotes have a remarkably well-conserved apparatus for the trafficking of proteins between intracellular compartments and delivery to their target organelles. This apparatus comprises the secretory (or 'protein export') pathway, which is responsible for the proper processing and delivery of proteins and lipids, and is essential for the derivation and maintenance of those organelles. Protein transport between intracellular compartments is mediated by carrier vesicles that bud from one organelle and fuse selectively with another. Therefore, organelle-specific trafficking of vesicles requires specialized proteins that regulate vesicle transport, docking and fusion. These proteins are generically termed SNAREs and comprise evolutionarily conserved families of membrane-associated proteins (i.e. the synaptobrevin/VAMP, syntaxin and SNAP-25 families) which mediate membrane fusion. SNAREs act at all levels of the secretory pathway, but individual family members tend to be compartment-specific and, thus, are thought to contribute to the specificity of docking and fusion events. In this review, we describe the different SNARE families which function in exocytosis, as well as discuss the role of possible negative regulators (e.g. 'SNARE-masters') in mediating events leading to membrane fusion. A model to illustrate the dynamic cycling of SNAREs between fusion-incompetent and fusion-competent states, called the SNARE cycle, is presented.
1998
-
(1998) Journal of Cell Biology. 143, 5, p. 1167-1182 Abstract
Members of the synaptobrevin/VAMP family of v-SNAREs are thought to be essential for vesicle docking and exocytosis in both lower and higher eukaryotes. Here, we describe yeast mutants that appear to bypass the known v-SNARE requirement in secretion. Recessive mutations in either VBM1 or VBM2, which encode related ER-localized membrane proteins, allow yeast to grow normally and secrete in the absence of Snc v-SNAREs. These mutants show selective alterations in protein transport, resulting in the differential trafficking and secretion of certain protein cargo. Yet, processing of the vacuolar marker, carboxypeptidase Y, and the secreted protein, invertase, appear normal in these mutants indicating that general protein trafficking early in the pathway is unaffected. Interestingly, VBM1 and VBM2 are allelic to ELO3 and ELO2, two genes that have been shown recently to mediate the elongation of very long chain fatty acids and subsequent ceramide and inositol sphingolipid synthesis. Thus, the v-SNARE requirement in constitutive exocytosis is abrogated by mutations in early components of the secretory pathway that act at the level of lipid synthesis to affect the ability of secretory vesicles to sort and deliver protein cargo.
-
Vesicular trafficking on the late secretory pathway of the budding yeast, S. cerevisiae(1998) Cellular and Molecular Mechanisms of Toxin Action: Secretory Systems. p. 81-108 Abstract
1997
-
(1997) Journal of Biological Chemistry. 272, 26, p. 16591-16598 Abstract
We are studying yeast homologs of the synaptobrevin/VAMP family of vesicle-associated membrane proteins, which act as vesicular compartment- soluble N-ethylmaleimide-sensitive factor attachment protein receptors (v- SNAREs) in cells having a capacity for stimulus-coupled secretion, as well as in other cell types. The yeast homologs, Snc1 and Snc2, localize to secretory vesicles and are required for normal bulk secretion in Saccharomyces cerevisiae. Here we have used Snc deletion mutants and chimetic Snc-VAMP proteins to demonstrate that these v-SNAREs can be dissected into regions that are either indispensable or dispensable for exocytic function in vivo. We have found that a region encompassing two predicted amphipathic α- helices (helix 1 and helix 2) (residues 32-85), which are thought to form coiled-coil structures, is essential for conferring exocytosis in yeast. Deletions in either the helix 1 or helix 2 segments result in a complete loss in the ability of the protein to confer secretion competence to snc cells and to interact genetically with components of the proposed fusion complex: the Sec9 and Sso2 t-SNAREs and the Sec17 α-SNAP homolog. In contrast, deletions in either the variable (residues 2-27) or putative intravesicnlar (residues 115-117) regions have no deleterious effect upon v-SNARE function. This makes it unlikely that sequences in either the amino or carboxyl terminus act in an exocytic capacity. Along with additional studies utilizing chimeric Snc-VAMP proteins, we suggest that although the Snc and synaptobrevin/VAMP proteins have evolved to mediate vastly different exocytic programs, their structural requirements and actions have remained remarkably well-conserved in evolution.
-
(1997) Journal of Biological Chemistry. 272, 1, p. 36-39 Abstract
We have previously reported the identification and partial characterization of a gene encoding a phospholipase D activity (PLD1) in the yeast, Saccharomyces cerevisiae. Here we report the existence of a second phospholipase D activity, designated PLD2, in yeast cells bearing disruption at the PLD1 locus. PLD2 is a Ca2+-dependent enzyme which preferentially utilizes phosphatidylethanolamine over phosphatidylcholine as a substrate. In contrast to PLD1, the activity of PLD2 is insensitive to phosphatidylinositol 4,5-bisphosphate, and the enzyme is incapable of catalyzing the transphosphatidylation reaction with short chain alcohols as acceptors. Subcellular fractionation shows that PLD2 localizes mainly to the cytosol, but could also be detected in the particulate fraction. Thus, the biochemical properties of PLD2 appear to be substantially different from those of PLD1. PLD2 activity is significantly and transiently elevated upon exit of wild type yeast cells from stationary phase, suggesting that it may play a role in the initiation of mitotic cell division in yeast. In view of the significantly different properties of PLD1 and PLD2, and because the yeast genome contains PLD1 as the sole member of the recently defined PLD gene family, it may be concluded that PLD2 is structurally unrelated to PLD1. Thus, the novel PLD2 activity described herein is likely to represent the first identified member of a new PLD gene family.
1996
-
(1996) Journal of Biological Chemistry. 271, 5, p. 2361-2364 Abstract
We have identified an open reading frame on chromosome XI of the yeast, Saccharomyces cerevisiae, as encoding a protein with phospholipase D (PLD) activity. We have named this open reading frame, PLD1, and show that yeast bearing a disruption in this gene are unable to catalyze the hydrolysis of phosphatidylcholine. PLD1 encodes a hypothetical protein of 1683 amino acids and has a predicted molecular mass of 195 kDa. Yeast bearing disruptions at the PLD1 locus are morphologically normal and grow vegetatively like wild- type cells. In contrast, homozygous Δpld1 diploid cells are unable to sporulate and do not produce asci under conditions that induce meiosis and sporulation in wild-type cells. Thus, PLD1 is likely to be essential for the meiotic cycle in yeast cells. This is the first identification of a eukaryotic, non-plant, phosphatidylcholine-hydrolyzing phospholipase D gene. Because the biological role of PLD is not well understood, we expect that Δpld1 yeast will become a useful tool for the characterization of PLD functions as well as for the identification of mammalian PLD homologs.
-
(1996) Journal of Biological Chemistry. 271, 30, p. 18243-18252 Abstract
The yeast adenylyl cyclase-associated protein, CAP, was identified as a component of the RAS-activated cyclase complex. CAP consists of two functional domains separated by a proline-rich region. One domain, which localizes to the amine terminus, mediates HAS signaling through adenylyl cyclase, while a domain at the carboxyl terminus is involved in the regulation of cell growth and morphogenesis. Recently, the carboxyl terminus of yeast CAP was shown to sequester actin, but whether this function has been conserved, and is the sole function of this domain, is unclear. Here, we demonstrate that the carboxyl-terminal domains of CAP and CAP homologs have two separate functions. We show that carboxyl-terminals of both yeast CAP and a mammalian CAP homolog, MCH1, bind to actin. We also show that this domain contains a signal for dimerization, allowing both CAP and MCH1 to form homodimers and heterodimers. The properties of actin binding and dimerization are mediated by separate regions on the carboxyl terminus; the last 27 amine acids of CAP being critical for actin binding. Finally, we present evidence that links a segment of the proline-rich region of CAP to its localization in yeast. Together, these results suggest that all three domains of CAP proteins are functional.
1995
-
(1995) Proceedings of the National Academy of Sciences of the United States of America. 92, 13, p. 5987-5991 Abstract
Yeast possess two homologs of the synaptobrevin family of vesicle- associated membrane proteins that function in membrane recognition and vesicle fusion. Yeast proteins Snc1 and Snc2 localize to secretory vesicles and are required for constitutive exocytosis. They also form a physical complex with a plasma membrane protein, Sec9, which is necessary for vesicle docking and fusion to occur in vivo. Formation of this molecular complex, as a prerequisite for vesicle fusion, appears to have been conserved evolutionarily. Here we demonstrate that Snc proteins undergo a single posttranslational modification with the addition of a palmitate moiety to Cys-95 in Snc1. Modification of Cys-95 (which is located proximal to the transmembrane domain) is rapid, occurs in the endoplasmic reticulum, and is long-lasting. Mutation of Cys-95 to Ser-95 blocks palmitoylation and appears to affect Snc protein stability. This provides evidence that synaptobrevin- like proteins are modified posttranslationally, and we predict that fatty acylation may be common to those found in higher eukaryotes.
1994
-
(1994) The journal of Biological chemistry. 269, 38, p. 23391-23394 Abstract
Yeast possess two homologs of the synaptobrevin family of vesicle-associated proteins that are proposed to be involved in membrane recognition and to act as receptors for components of the fusion machinery in neurons. We have previously described the yeast homologs, Snc1 and Snc2, and demonstrated that they localize to secretory vesicles and are required for normal secretion. Yeast lacking Snc protein expression accumulate post-Golgi transport vesicles that contain secretory proteins. Therefore, Snc proteins are essential for the fusion of carrier vesicles with the plasma membrane, and this property appears to have been conserved in evolution. We have now examined whether Snc proteins interact with other components of the late secretory pathway in yeast. Here we show that Snc proteins form a tight genetic and physical interaction with a plasma membrane protein, Sec9. Sec9 is the yeast equivalent of SNAP-25, a second receptor protein from neurons that has been shown to interact with synaptobrevin. We suggest, then, that recognition of the plasma membrane by secretory vesicles may involve the formation of a Snc-Sec9 complex and that this interaction has evolved as a fundamental step in secretory processes.
-
SNC1 and SNC2(1994) A Guidebook to the Secretory Pathway. Rothblatt J. & Novick P.(eds.). p. 172-173 Abstract
[Book Abstract] The late 1970s and early 1980s saw a boom in research establishing molecular details related to secretory protein modifications such as signal peptide processing, proenzyme proteolytic maturation, glycosylation, and oligosaccharide modification. This timely new resource compiles what is known about the secretory pathway. The book charts how much of the current progress in the field derives from the development of cell-free systems that reproduce individual transport steps, the identification of proteins associated with vesicular carriers, and the isolation of mutants and characterization of gene products that mediate protein translocation and secretion. Guidebook to the Secretory Pathway will be invaluable to researchers and students of cell and molecular biology.
1993
-
(1993) Cell. 74, 5, p. 855-861 Abstract
The yeast S. cerevisiae possesses two genes, SNC1 and SNC2, that encode homologs of vertebrate synaptic vesicle-associated membrane proteins, also known as synaptobrevins. Here we describe the cloning of SNC2 and demonstrate that yeast lacking both SNC genes are deficient in normal bulk secretion, accumulate large numbers of post-Golgi vesicles, and display a variety of conditional lethal phenotypes. In addition, we show that yeast Snc proteins localize to post-Golgi transport vesicles that accumulate in a late-acting sec mutant. Our findings clearly place the Snc proteins on the late secretory pathway in S. cerevisiae and provide direct in vivo evidence that synaptobrevin-like proteins mediate the targeting and transport of secretory proteins.
-
1992
-
Genetic and biochemical analysis of the adenylyl cyclase-associated protein, cap, in Schizosaccharomyces pombe(1992) Molecular Biology of the Cell. 3, 2, p. 167-180 Abstract
We have identified, cloned, and studied a gene, cap, encoding a protein that is associated with adenylyl cyclase in the fission yeast Schizosaccharomyces pombe. This protein shares significant sequence homology with the adenylyl cyclase-associated CAP protein in the yeast Saccharomyces cerevisiae. CAP is a bifunctional protein; the N-terminal domain appears to be involved in cellular responsiveness to RAS, whereas loss of the C-terminal portion is associated with morphological and nutritional defects. S. pombe cap can suppress phenotypes associated with deletion of the C-terminal CAP domain in S. cerevisiae but does not suppress phenotypes associated with deletion of the N-terminal domain. Analysis of cap disruptants also mapped the function of cap to two domains. The functional loss of the C-terminal region of S. pombe cap results in abnormal cellular morphology, slow growth, and failure to grow at 37°C. Increases in mating and sporulation were observed when the entire gene was disrupted. Overproduction of both cap and adenylyl cyclase results in highly elongated large cells that are sterile and have measurably higher levels of adenylyl cyclase activity. Our results indicate that cap is required for the proper function of S. pombe adenylyl cyclase but that the C-terminal domain of cap has other functions that are shared with the C-terminal domain of S. cerevisiae CAP.
-
SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: Genetic interactions with the RAS and CAP genes(1992) Proceedings of the National Academy of Sciences of the United States of America. 89, 10, p. 4338-4342 Abstract
SNC1, a gene from the yeast Saccharomyces cerevisiae, encodes a homolog of vertebrate synaptic vesicle-associated membrane proteins (VAMPs) or synaptobrevins. SNC1 was isolated by its ability to suppress the loss of CAP function in S. cerevisiae strains possessing an activated allele of RAS2. CAP is a component of the RAS-responsive S. cerevisiae adenylyl cyclase complex. The N-terminal domain of CAP is required for full cellular responsiveness to activated RAS proteins. The C-terminal domain of CAP is required for normal cellular morphology and responsiveness to nutrient extremes. Multicopy plasmids expressing SNC1 suppress only the loss of the C-terminal functions of CAP and only in the presence of activated RAS2.Errata: As a result of a clerical oversight, an error in the numbering
of the nucleotides as well as a missing nucleotide in the intron sequence of SNCI
occurred.
1991
-
(1991) Cell. 66, 3, p. 497-505 Abstract
CAP is a component of the S. cerevisiae adenylyl cyclase complex. The N-terminal domain is required for cellular RAS responsiveness. Loss of the C-terminal domain is associated with morphological and nutritional defects. Here we report that cap cells bud randomly and are defective in actin distribution. The morphological and nutritional defects associated with loss of the CAP C-terminal domain are suppressed by over-expression of PFY, the gene encoding profilin, an actin- and polyphosphoinositide-binding protein. The phenotype of cells lacking PFY resembles that of cells lacking the CAP C-terminal domain. Study of mutated yeast profilins and profilins from Acanthamoeba suggests that the ability of profilin to suppress cap cells is dependent upon a property other than, or in addition to, its ability to bind actin. This property may be its ability to bind polyphosphoinositides. We propose that CAP and profilin provide a link between growth signals and remodeling of the cellular cytoskeleton.
-
(1991) Molecular and Cellular Biology. 11, 3, p. 1248-1257 Abstract
CAP, a protein from Saccharomyces cerevisiae that copurifies with adenylyl cyclase, appears to be required for yeast cells to be fully responsive to RAS proteins. CAP also appears to be required for normal cell morphology and responsiveness to nutrient deprivation and excess. We describe here a molecular and phenotypic analysis of the CAP protein. The N-terminal domain is necessary and sufficient for cellular response to activated RAS protein, while the C-terminal domain is necessary and sufficient for normal cellular morphology and responses to nutrient extremes. Thus, CAP is a novel example of a bifunctional component involved in the regulation of diverse signal transduction pathways.
1990
-
Cloning and characterization of CAP, the Saccharomyces cerevisiae gene encoding the 70 kDa adenylyl cyclase-associated protein(1990) Cell. 61, p. 319-327 Abstract
1989
-
(1989) European Journal of Pharmacology - Molecular Pharmacology Section. 172, 1, p. 29-39 Abstract
We have examined the effects of a biologically active tumor promoting phorbol ester (phorbol 12-myristate, 13-acetate (PMA)) which activates protein kinase C (PKC) on melanotropin receptor function and cell growth in the M2R mouse melanoma cell clone. Treatment of M2R cells with PMA resulted in a significant loss of β-MSH binding. The effect was both time- and concentration-dependent. The inhibition of β-MSH binding resulted from a decrease (> 85%) in active membranal receptors available on the external cell surface and not from either enhanced internalization or change in the binding affinity. Agonist-stimulated cyclic AMP accumulation was profoundly increased in a non-selective manner following short-term incubation (3 h) with PMA. This effect was completely reversed during long-term (72-96 h) incubation with the tumor promoting agent. Long-term culturing of M2R cells with PMA resulted in enhanced (+ 50%) proliferation of the melanoma cells. This enhancement was blocked by the addition of agents which stimulate the production of cAMP. Hence, phorbol esters are powerful growth promoters in transformed melanocytes and our findings indicate that the effects of melanotropins are selectively impaired during the process of growth promotion.
1988
-
(1988) Endocrinology. 123, 4, p. 1792-1797 Abstract
In this study, two melanotropin binding proteins from M2R melanoma cells have been identified based on the photochemical cross-linking of [125I]iodinated porcine β-MSH ([125I]iodo-β-MSH) to melanoma cell membranes, using iV-hydroxysuccinimidyl- azidobenzoate. Autoradiography of photoaffinity- labeled M2R membrane protein, after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, revealed the specific labeling of two separate bands with an apparent molecular mass of 43 and 46 kilodaltons, respectively. Photoaffinity labeling of both bands was of near-equal intensity and could be inhibited, in a dose-dependent manner, by the addition of unlabeled β- MSH before photolysis. In addition, agents known to inhibit the binding of β-MSH to its cellular receptor, such as EGTA, GTP, guanosine 5-O-(3-thio)triphosphate, and a synthetic analog of the calmodulin-binding domain of myosin light chain kinase- M5, were all found to specifically inhibit the labeling of these two protein bands by the azido derivative of [125I]iodo-β-MSH. In contrast, addition of a nonrelated peptide, vasoactive intestinal peptide, had no effect upon the labeling of these melanotropin- binding proteins. On the basis of these results we suggest that the two proteins may function as the binding domain(s) of the cellular receptor for melanotropins, or may represent entire receptor moieties themselves.
-
A synthetic analog of the calmodulin-binding domain of myosin light chain kinase inhibits melanotropin receptor function and activation of adenylate cyclase(1988) Journal of Biological Chemistry. 263, 15, p. 7073-7078 Abstract
In this study a synthetic analog of the calmodulin-binding domain of myosin light chain kinase, a 17-amino-acid peptide (M5) was used to examine the possible role of calmodulin in melanotropin receptor function. Binding of beta-melanocyte-stimulating hormone to its membrane receptor and subsequent stimulation of adenylate cyclase (AC) were found to be specifically inhibited by M5 in a dose-dependent and noncompetitive manner, both in intact M2R melanoma cells and in a plasma membrane preparation derived thereof. Half-maximal inhibition of both hormone binding and melanotropin-sensitive AC activity was shown to occur at approximately 1 microM M5. In contrast, stimulation of AC by prostaglandin E1, guanosine 5'-O-(3-thio)triphosphate, forskolin, and unstimulated enzyme activity were unaffected by the presence of M5, under the same assay conditions. Furthermore, addition of a molar excess of calmodulin to the AC assay completely abolished the inhibitory effects of the peptide on melanotropin-stimulated AC activity. Other peptides of similar size, which bind to calmodulin with low affinity (e.g. glucagon, somatostatin, and vasoactive intestinal peptide), were shown to be totally ineffective in inhibiting melanotropin-sensitive AC. These findings, along with those shown previously for other antagonists of calmodulin, suggest a role for an M5-binding protein, as of yet unidentified, involved in the regulation of the melanotropin receptor function.
-
(1988) Hormones and cancer 3. Bresciani F.(eds.). p. 164-167 (trueProgress in Cancer Research and Therapy). Abstract
1987
-
(1987) Endocrinology. 121, 5, p. 1766-1772 Abstract
Melanotropin (MSH) receptor activity in the M2R mouse melanoma cell line is tightly controlled by calcium by an unknown mechanism. The possibility that calcium regulation is mediated by calmodulin or a calmodulin-related calcium binding protein has been addressed in this report by studying the effects of two known calmodulin antagonists, fluphenazine and melittin, on MSH receptor function. Stimulation of adenylate cyclase (AC) in M2R plasma membranes by βMSH was strongly inhibited by both antagonists. The concentrations of fluphenazine and melittin yielding half-maximal inhibition (IC50) of AC were 16 nM and 2.4 μM, respectively. Both fluphenazine and melittin also inhibit prostaglandin E1-, GTPÎ3S, and forskolin-stimulated AC activity, as well as that of unstimulated enzyme, although inhibition is shown to occur at significantly higher concentrations of antagonist. We have shown that the calcium-dependent rate-limiting step in MSH stimulation of adenylate cyclase, that of hormone binding, is strongly inhibited by these antagonists at concentrations identical to, if not lower than, those required for the inhibition of AC activity (fluphenazine- IC50, 14 μM; melittin-IC60, 0.7 μM). The actions of these antagonists, furthermore, appear to be calcium insensitive, as melittin affects the stability of both the high affinity (calcium containing) and low affinity (calcium depleted) receptor-MSH complexes. The sensitivity of the MSH receptor to inhibition by calmodulin antagonists resembles that described for purified calmodulin-sensitive enzyme systems, which suggests a possible role for calmodulin in MSH receptor function. Among peptide hormone receptors, this effect by calmodulin antagonists appears to be unique for the MSH receptor.
-
-
(1987) Molecular Pharmacology. 31, 1, p. 81-88 Abstract
Binding of β-melanotropin (β-MSH) and subsequent activation of adenylate cyclase in the M2R mouse melanoma cell line is strongly dependent on the concentration of extracellular free calcium. This effect can be demonstrated both in the intact cell and in a plasma membrane preparation derived therefrom, using an EGTA buffer system. In contrast, stimulation of adenylate cyclase by prostaglandin E1, forskolin, or guanosine 5'-O-(2-thiotriphosphate) is calcium insensitive. It is shown that calcium increases the binding affinity of β-MSH for its receptor by a factor of 20 (from 400 nM to 20 nM) without affecting maximal hormone binding. At supersaturating concentrations of β-MSH (> 200 nM) binding gradually becomes calcium independent. Hormone-receptor complexes formed in the presence of calcium dissociated rapidly (≤ 2 min) and reversibly upon the elimination of calcium by excess EGTA. Among nine divalent metal cations tested, calcium was found to be the most effective in facilitating hormone binding. Whereas calcium promotes β-MSH binding, GTP and its stable analogs lead to a reduction in both maximal binding (65%) and affinity (2-fold). These effects are calcium independent, suggesting that the reciprocal control of β-MSH binding by calcium and guanosine nucleotides is mediated by two separate and independent mechanisms.
1986
-
(1986) Molecular and Cellular Endocrinology. 46, 2, p. 137-147 Abstract
We have examined adenylate cyclase (AC) in the M2R melanoma cell line, a novel clone of transplantable B16 melanoma cells. It has been found that activity of this enzyme is highly responsive to β-melanotropin (β-MSH) and other hormones possessing melanotropic activity (e.g., α-melanotropin (α-MSH) and adrenocorticotrophic hormone (ACTH1/224)). β-MSH stimulation of adenylate cyclase, both in the intact cell and in a plasma membrane-enriched fraction derived thereof, was shown to be saturable and dose-dependent. In addition, prostaglandin E1 (PGE1) was found to be a potent stimulator of AC activity in these cells. Hormone stimulation of enzyme activity in the intact cell was strongly potentiated by forskolin which not only enhanced maximal AC activity 3-fold, but lowered by 40-fold the concentration of β-MSH required for half-maximal stimulation. Using biologically active [125I]iodo-β-MSH prepared in our laboratory we have examined the specificity of β-MSH binding to its receptor in both intact M2R cells and plasma membranes derived thereof. Among a series of hormones tested only α-MSH and ACTH1-24 competed with [125I]iodo-β-MSH for binding to the melanotropin receptor in accordance with the results obtained with AC. In contrast to the strong effect on cyclic 3',5'-adenosine monophosphate (cAMP) accumulation in M2R cells forskolin has no effect on [125I]iodo-β-MSH binding. It appears that the kinetic properties of β-MSH binding and β-MSH stimulation of adenylate cyclase activity are essentially identical, the half-maximal effects of which are demonstrated at approximately 20 nM β-MSH.
1984
-
Biosynthesis of steroids by the ovarian component of Sparus aurata(1984) European Mariculture Society: Research on Aquaculture. p. 139-149 Abstract
Biosynthesis of steroids by the ovarian component of Sparus aurata