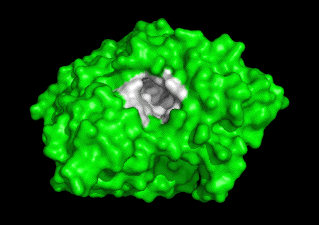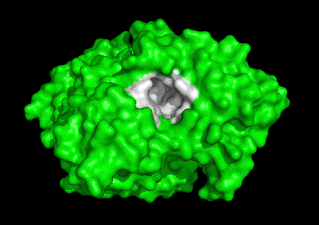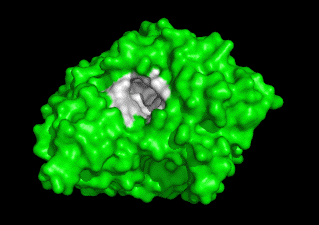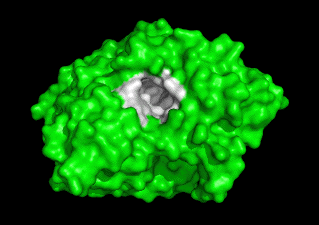
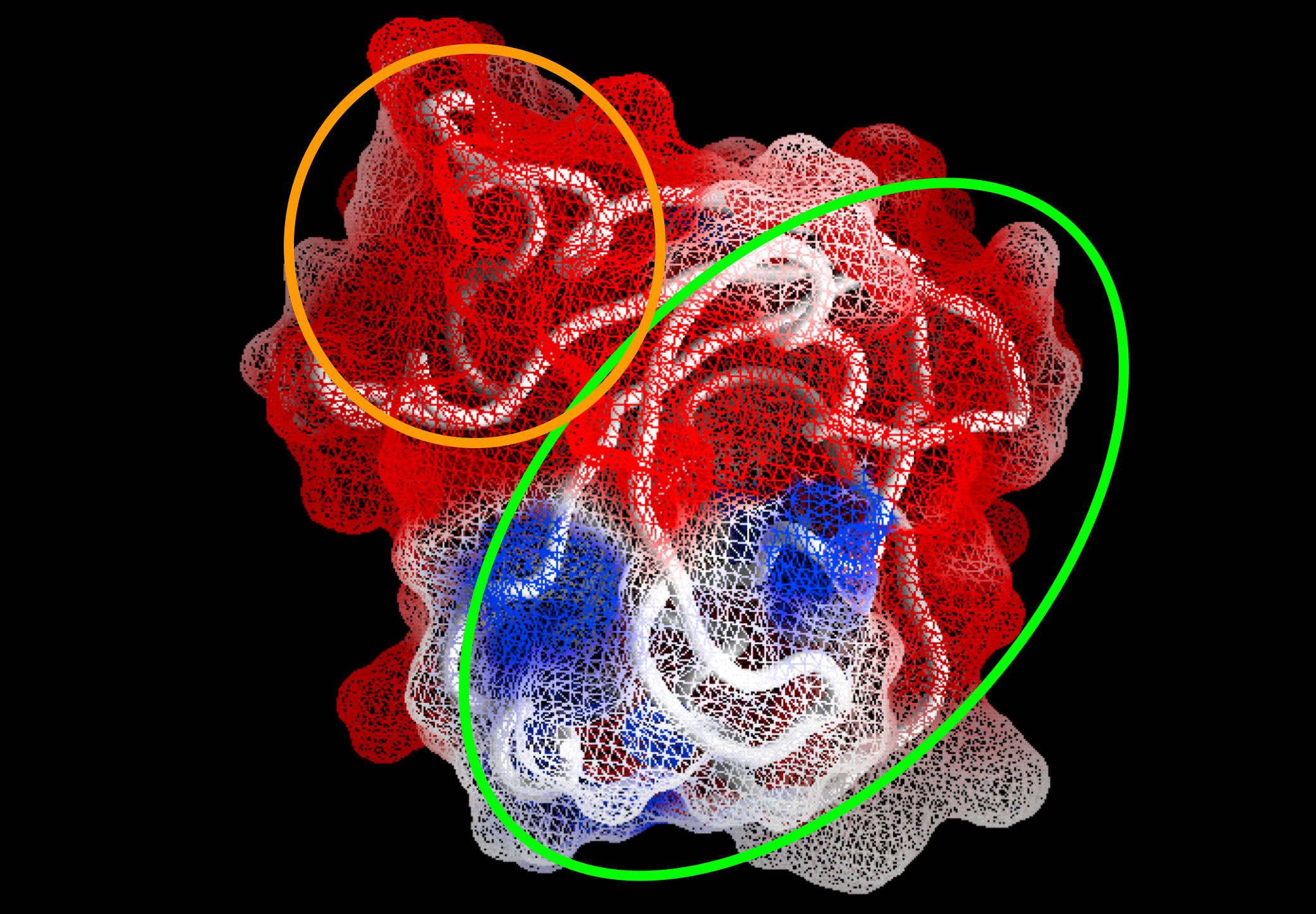
Together with Prof. Israel Silman, we are studying the 3D structure/function of nervous system proteins, such as acetylcholinesterase (AChE), cholinesterase-like adhesion molecules (CLAMs), β-glucosidase, β-secretase, paraoxonase and intrinsically disordered proteins. Although our research is curiosity driven, the findings may impact the treatment of neurological disorders, including Alzheimer's Disease & autism.
We've helped to create and continue to develop:
- The Israel Structural Proteomics Center (ISPC)
- Proteopedia, the collaborative, 3D encyclopaedia of biomacromolecules
For a podcast interview with Joel Sussman, see Meet the Scientists.