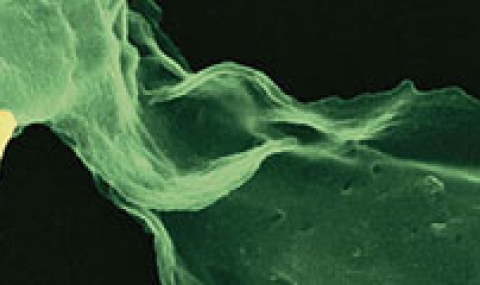
OUR APPROACH: Using comprehensive, quantitative, unbiased tools to analyse the molecular interactions that underlie distinct host-pathogen subpopulations and their impact on disease outcome.

The interface between immune cells and intracellular bacterial pathogens produces complex, diverse interactions. Within individual encounters, highly adaptable and dynamic host cells and bacteria vary in their responses, which can then underlie well-documented heterogeneous outcomes of infection. The challenge now is to break down multidimensional distributions that comprise this heterogeneity into informative readouts of population physiology and predictors of responses to infection.
Using genomic approaches that combine microscopy to follow infection outcome and single cell RNA-seq methods we aim to analyze the breadth heterogeneity as a strategy of the bacteria to optimize its survival during infection and of the host to eradicate the pathogen.
See: Avraham et. al. Cell 2015
Barczack, Avraham et. al., Plos Pathogens 2017