- Proteasomal degradation of IDPs
- Function of IDPs
- IDPs and metabolism
Intrinsically disordered proteins (IDPs)
It is well known that many proteins must fold into a specific conformation in order to perform their dedicated tasks. However, there is a class of proteins for which the disordered state is their natural state, and in fact this lack of defined structure enables them to perform many different functions. Many of these IDPs perform regulatory functions, and their lack of defined structure enables them to interact with a number of different partners.
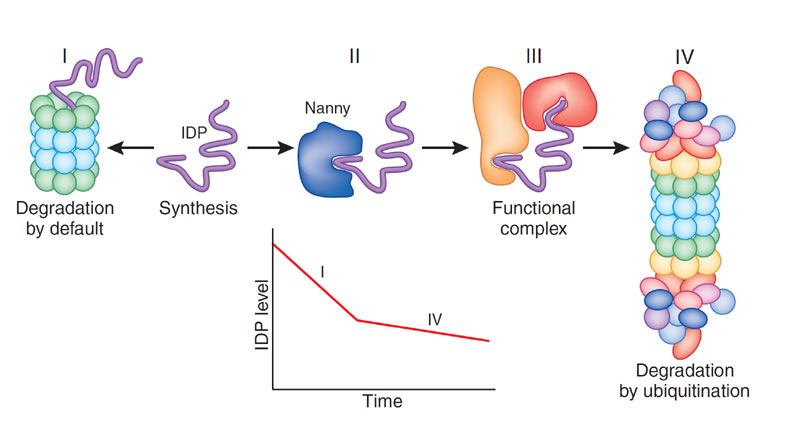
Proteasomal degradation of IDPs
The proteasome, a barrel-shaped protein-degrading complex, is used by the cell to regulate protein levels. Proteins tagged by ubiquitin are recognized and bound by the 19S cap of the 26S proteasome. The 19S cap also has de-ubiquitinating and unfolding activities, and the unfolded protein is fed into the narrow opening of the 20S catalytic core of the proteasome. Since IDPs are naturally unfolded, they are ready substrates for degradation by the 20S proteasomes, a process we call "degradation by default". The proteins can avoid degradation by binding to another protein or macromolecule, a "nanny". Our laboratory studies the regulation of IDP protein levels by default degradation and interaction with "nannies".
IDPs and metabolism
We found that IDP degradation by default via the 20S proteasomes is regulated by NADH and NQO1. In addition we revealed a new type of 26S proteasome that is ATP less but requires NADH. These findings couples IDP degradation with metabolic state of the cells.