What all the best-dressed proteins are wearing
Briefs
Just like clothes “make the man,” post-production chemical tags, like fashion accessories, make the protein. An entire wardrobe of protein modification tags— dubbed as such because they are added on after a protein has been made—helps the thousands of proteins in our bodies perform their versatile tasks. At the same time, these modifications make life difficult for scientists and physicians seeking to identify these vital molecules. Prof. Yifat Merbl’s lab in the Department of Systems Immunology has taken up the challenge of simultaneously detecting large numbers ofmodified proteins, together with all their accouterments.
Protein modification tags can make all the difference in how a protein functions—including its stability, location in the cell, and ability to bind to other molecules. Some tags might mark a diseased protein for destruction or determine how well another, such as a tumor suppressor, will do its job. Therefore, detecting and analyzing modified proteins is essential for the study of various body processes and diseases.
But with some 20,000 proteincoding genes in the human genome, each of which can come in four or five versions, there are approximately 100,000 possible protein sequences. Once a protein is created, it can be chemically modified by one or more of some 200 different tags. Multiply that number by the fact that these tags can attach to a protein molecule at multiple positions, and you get a virtually limitless number of possible tagging combinations and, consequently, of differently modified proteins.
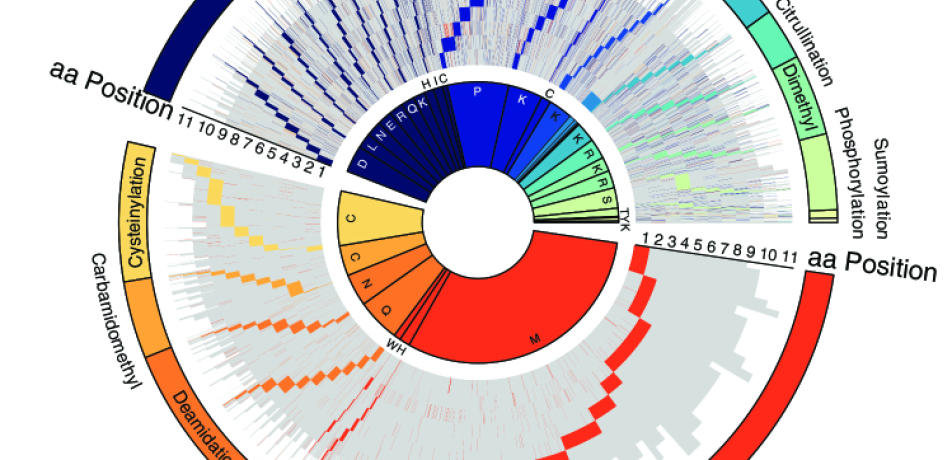
The Merbl lab has set itself the goal of creating a novel computational tool that can track down dozens of protein modification types in an unbiased manner. The Merbl lab’s search strategy—which they termed Protein Modification Integrated Search Engine, or PROMISE—looks for numerous alterations in parallel, rather than individually, as was done in the past. And PROMISE lives up to its name: It can identify some 30 modifications within six hours, whereas existing methods could identify only three at a time and took about two weeks to do it. The lab has published a paper describing PROMISE in Nature Biotechnology.
“PROMISE supplies us with new glasses, providing researchers with a broader picture of biological processes, and at a higher resolution than ever before,” Prof. Merbl says. “We can now use this tool to address unsolved biomedical problems by reexamining various kinds of existing datasets.”
In a preliminary test of PROMISE, the researchers sought to clarify whether and how protein modifications affect the immune system’s ability to detect tumors. The result is the most comprehensive atlas available of modified antigens characteristic of cancerous tumors. It includes a new “dictionary” showing how modifications occurring at specific sites in peptides may affect the immune system’s recognition of the cancerous cell.
This knowledge is critical because some of these modifications might explain how cancer cells “cloak” themselves and are then able to evade the T cells used in cancer immunotherapy. This understanding could in turn help researchers develop anti-cancer vaccines or T-cell therapies that target the modified antigen—should the modification be due to cancer— and improve the targeted attack against the tumor.
In future studies, PROMISE might also help reveal peptidemodification combinations that mislead immune cells into attacking healthy cells, causing autoimmune diseases. The tool might also provide new insights into neurodegenerative disorders and other diseases.
“Being able to quickly analyze protein modifications accurately can help us better diagnose numerous pathological processes, assess the response to drugs and ultimately develop new therapies,” Prof. Merbl says.
Yifat Merbl is supported by:
- Miel de Botton
- Dr. Gilbert S. Omenn and Martha A. Darling Weizmann Institute - Schneider Hospital Fund for Clinical Breakthroughs through Scientific Collaborations
- Sir John Ritblatt
- Dr. Barry Sherman Institute for Medicinal Chemistry