Research
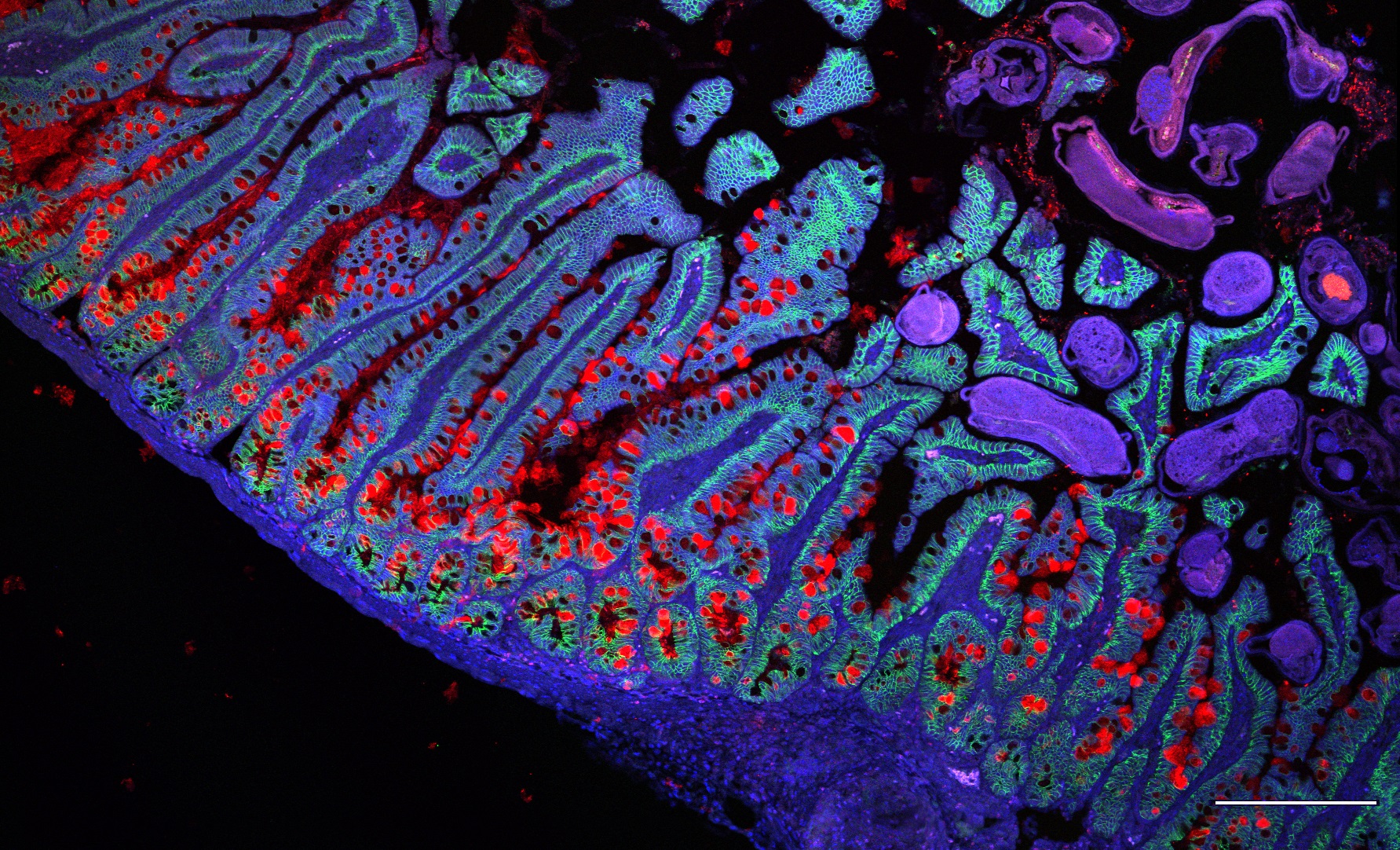
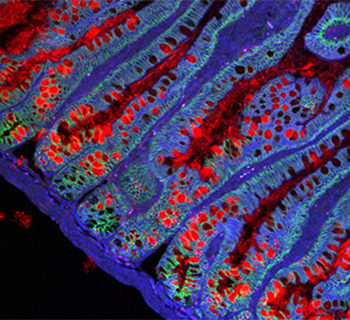
Our lab is utilizing experimental and computational methods to explore gut tissue homeostasis and pathologies. In this age of genomic approaches, it has now become feasible to characterize “normal” and diseased tissues in great detail. To understand the gut tissue physiology at higher resolution, we are leveraging cutting edge technologies such as single-cell genomics and traditional experimental methods.
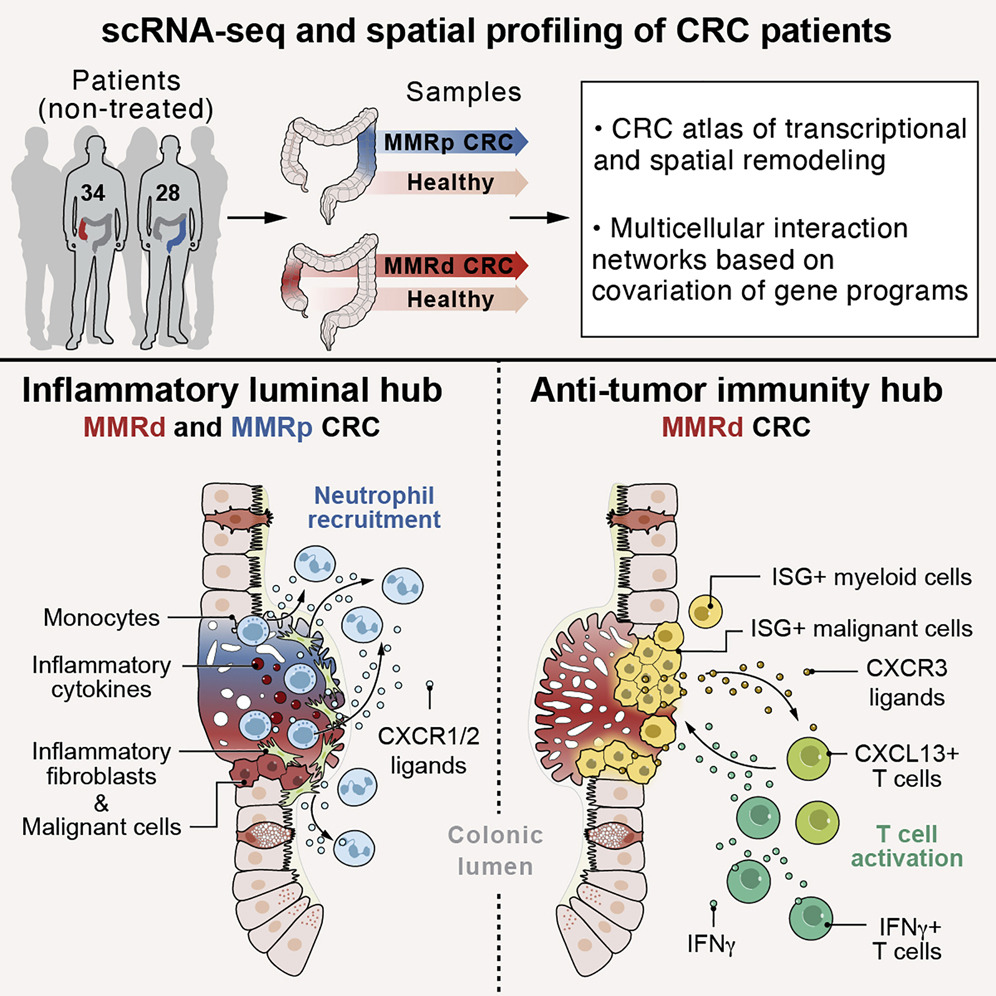
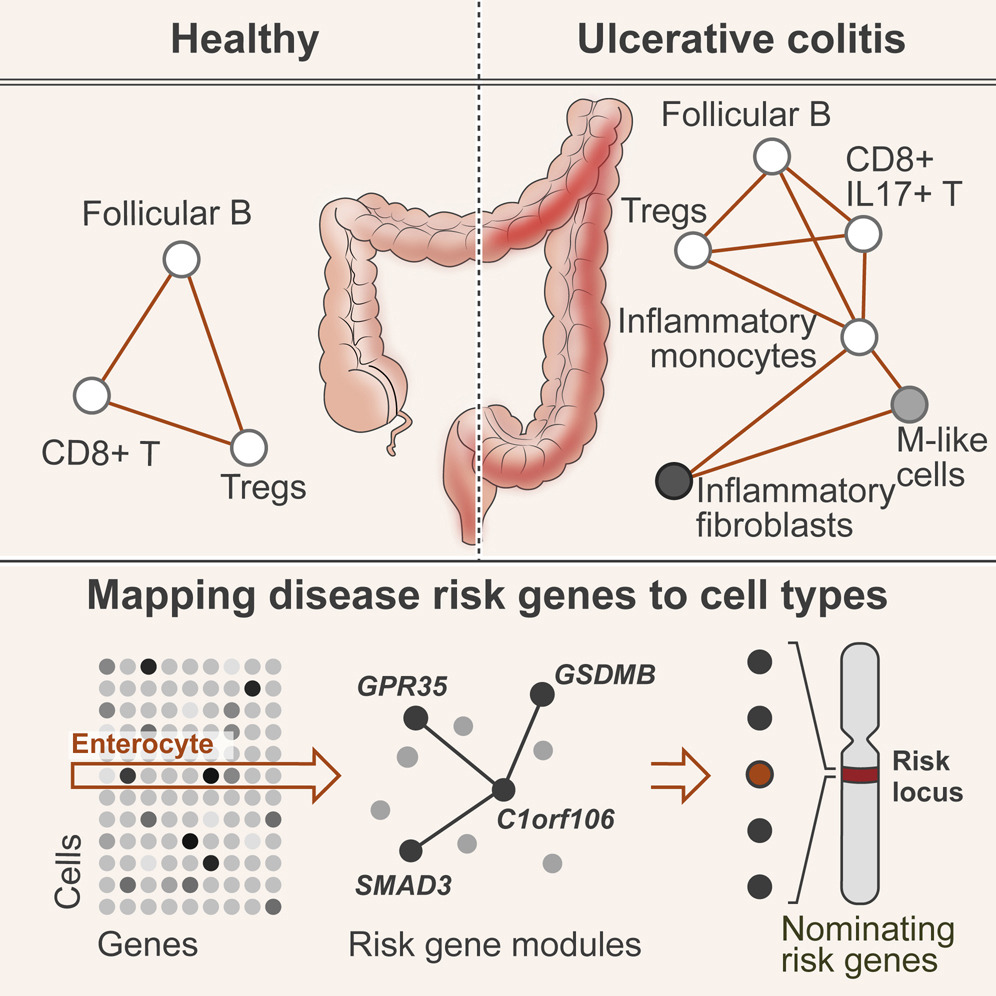
The gut is a complex ecosystem in which signals arriving from the outside world play a key role in shaping the mucosal surfaces and demanding multifactorial responses from diverse cell types that reside within the tissue. Disruption of this cellular equilibrium may lead to various tissue diseases, including food allergies, inflammatory bowel diseases, and cancer.
We aim to decipher the gut gene regulatory networks and explore their biological function by in vitro organoid systems or in vivo using mouse models and human samples. Our group is investigating the role of epithelial, intestinal stem cells, epithelial-immune interactions, inflammatory bowel disease (IBD) causal risk genes, and cancer processes in the intestines.
We focus on
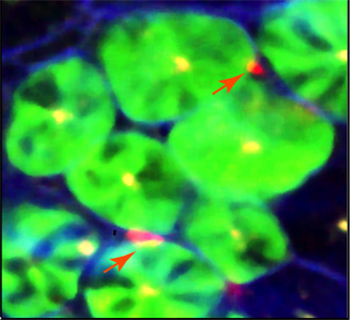
- Immune regulation of epithelial stem cells.
- Understanding the role of epithelial cells in shaping the mucosal immune cells.
- T helper cells' role in maintaining tissue homeostasis.
- Changes in cell-cell interactions upon different gut pathologies such as food allergies, IBD, and cancer.