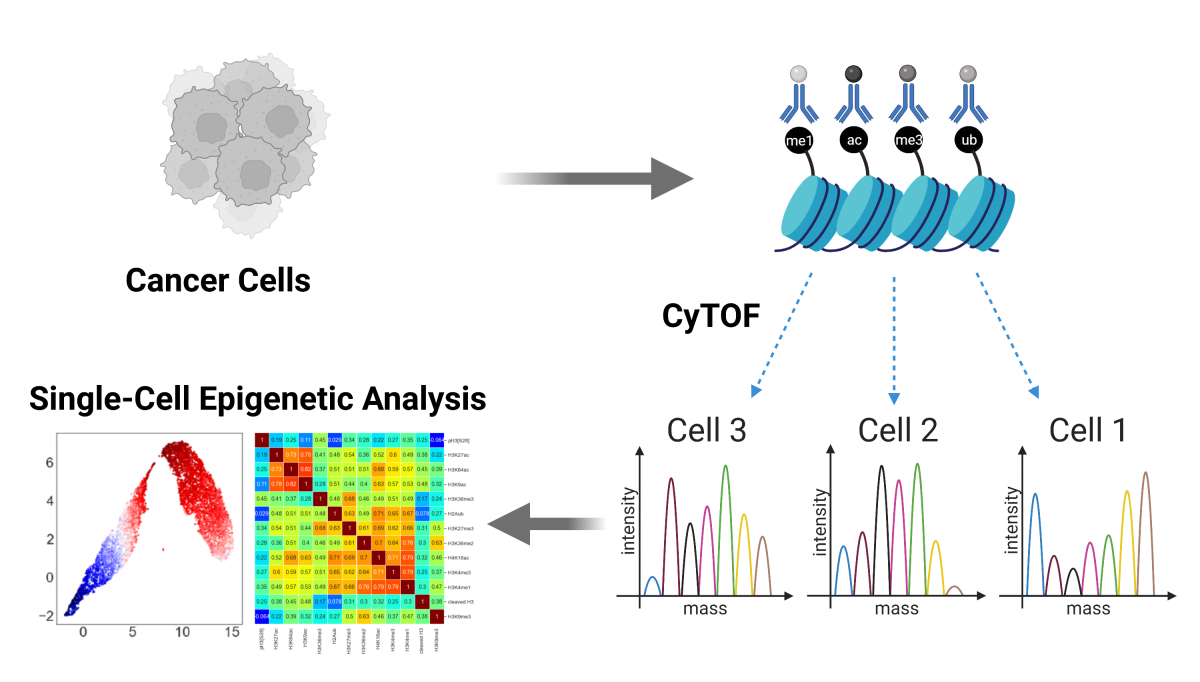
Cancer cells are highly heterogeneous, both at the transcriptional level and at their epigenetic state. While single cell RNA sequencing technologies have revolutionized our understanding of transcriptional heterogeneity, the plethora of histone post-translational modifications (PTMs) is still mainly analyzed using bulk methods. We adapted Cytometry by Time of Flight (CyTOF) to analyze a wide panel of histone modifications and chromatin regulators in various cancer models.
Analysis of primary tumor-derived lines of Diffused Intrinsic Pontine Glioma (DIPG), a fatal pediatric tumor harboring the H3-K27M oncohistone mutation, revealed two epigenetically distinct subpopulations, which differ in proliferation capacity as well as expression of stem-cell and differentiation markers. We strive to further explore the nature of this epigenetic heterogeneity; the molecular mechanisms that maintain it and the sensitivity of each subpopulation to drug treatments.