Single Cell Physiology
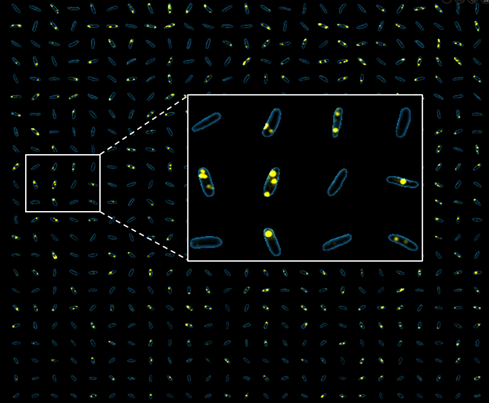
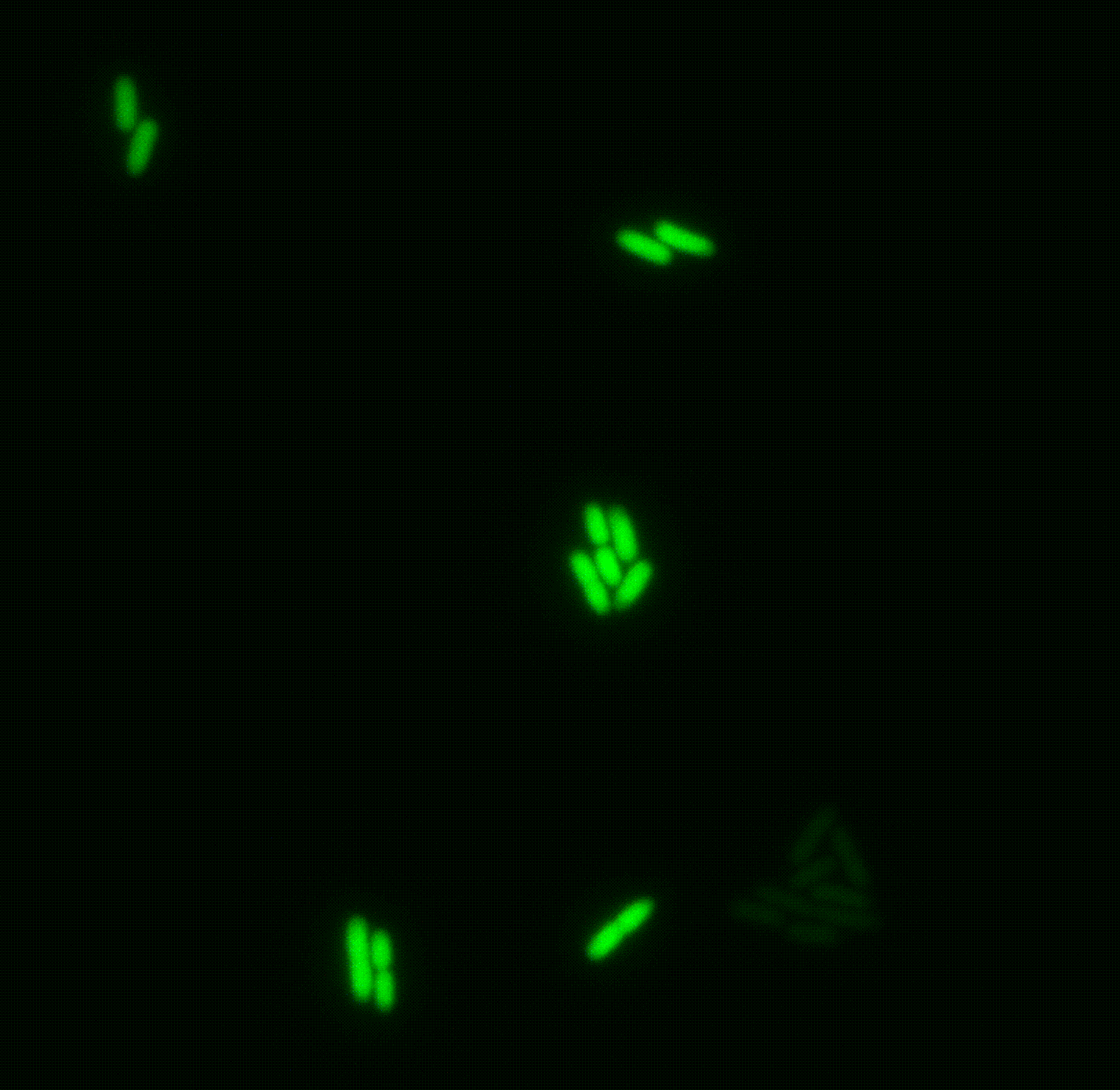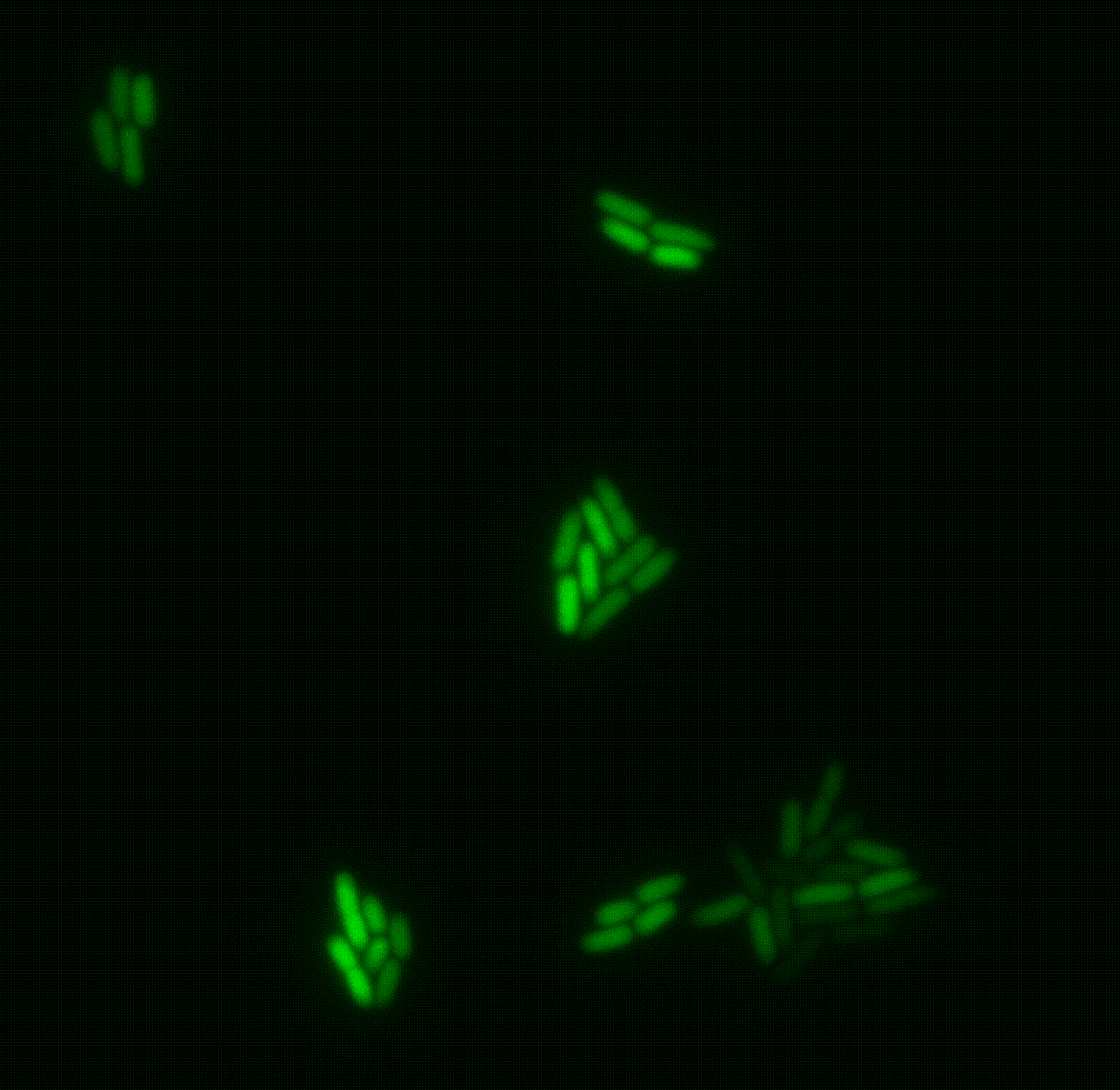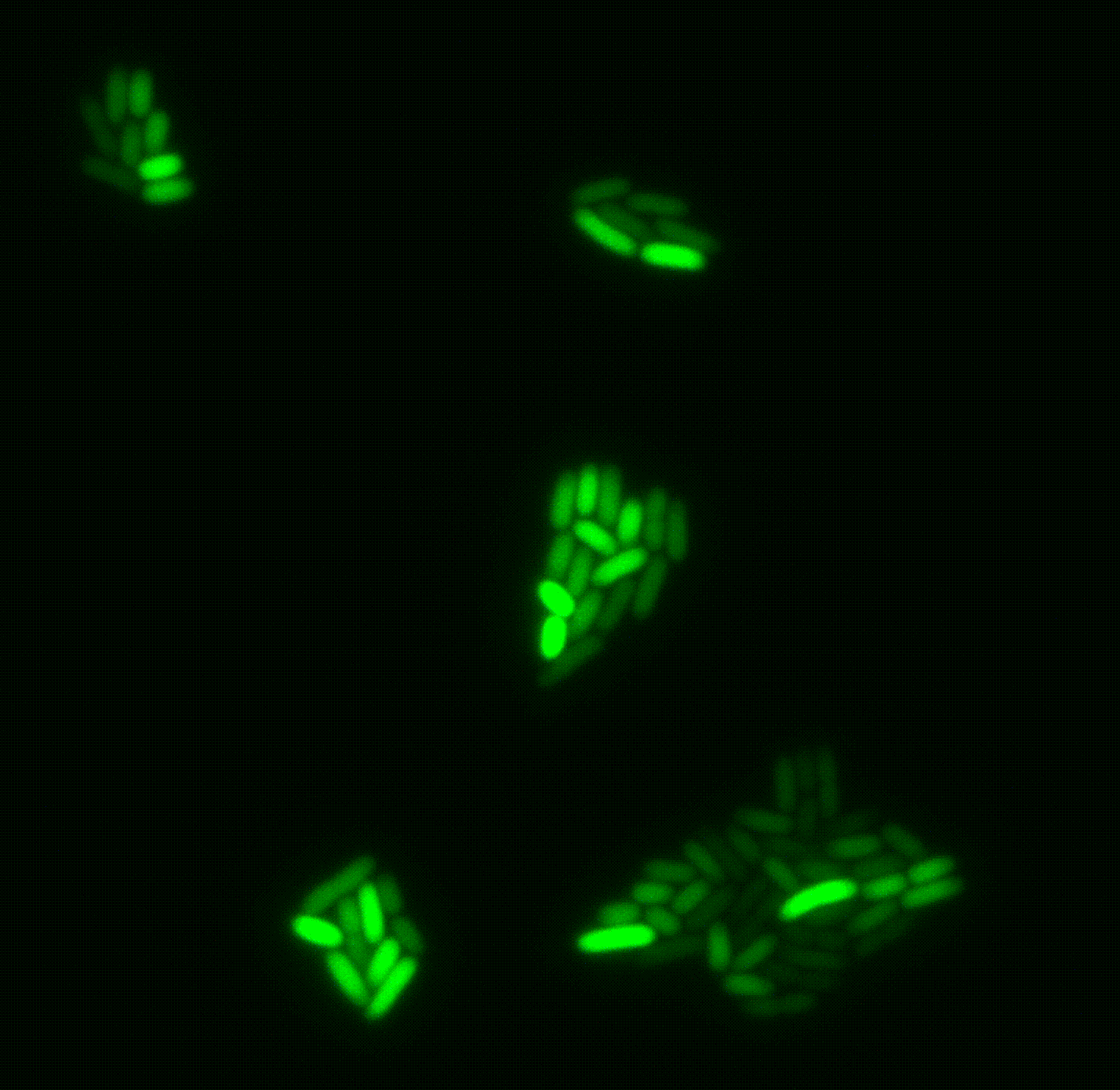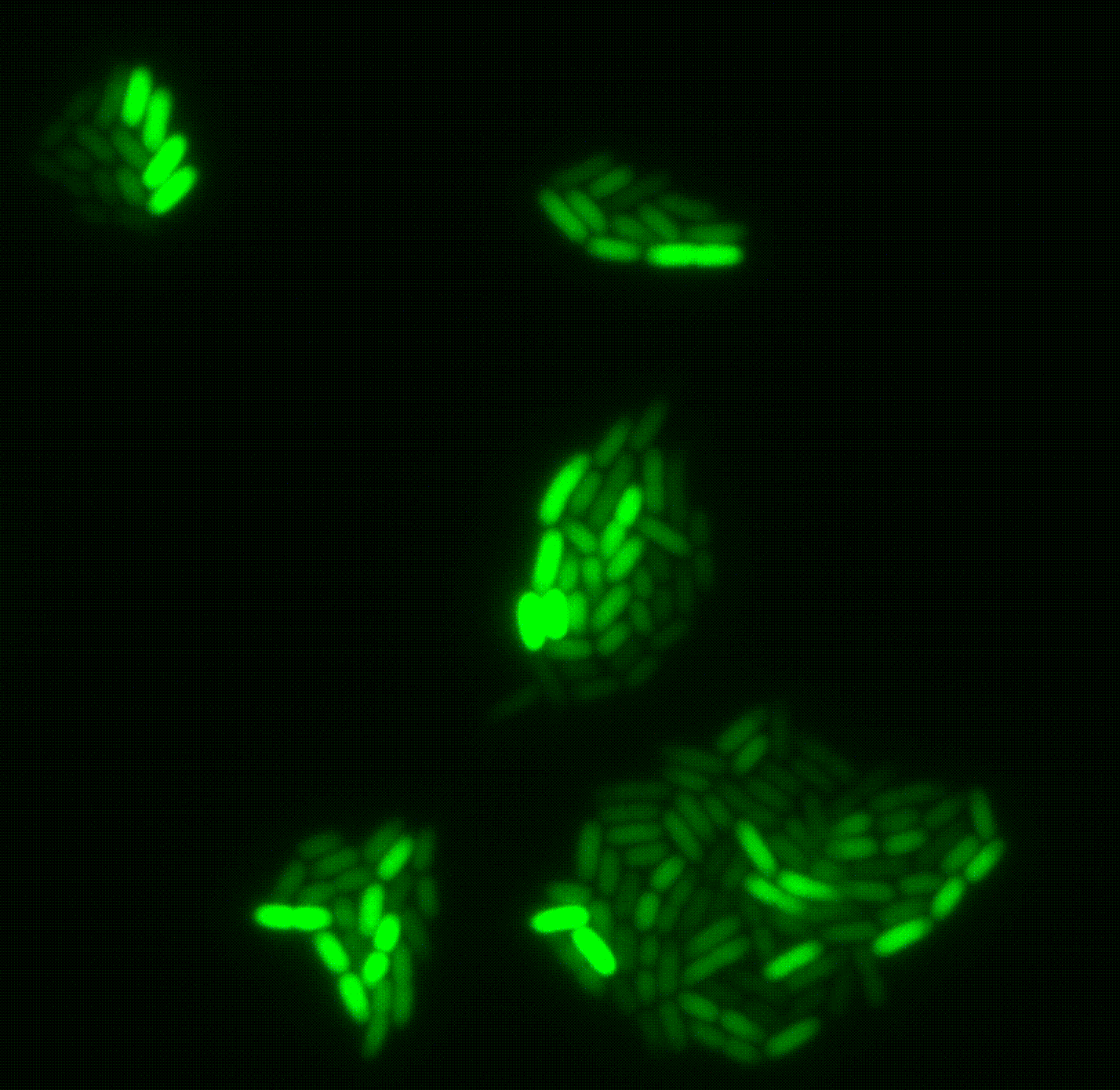
A clonal bacterial population may appear uniform, yet individual cells can differ markedly in their physiological state, even when grown in identical environments. These differences often arise from stochastic fluctuations (“noise”), but are frequently regulated so that only specific subpopulations express certain genes. Such phenotypic heterogeneity is critical for survival under acute stress, including antibiotic exposure. However, traditional bulk approaches average across millions of cells, obscuring this diversity and limiting its study.
We are developing new single-cell transcriptomics and live-imaging approaches to systematically uncover this heterogeneity with the aim of elucidating key features of bacterial physiology and life cycle. We focus on pathogenic bacteria, such as Pseudomonas and Salmonella.