Very recently, cellulosomes were revealed in the human gut, due to the sequencing of metagenomic samples and isolation and characterization of novel human gut bacterial species that grow on complex polysaccharide substrates.
Microcrystalline cellulose degrader
A cellulolytic fiber-degrading bacterium, Ruminococcus champanellensis, was isolated from human faecal samples, and its genome was recently sequenced. Bioinformatic analysis of the R. champanellensis genome revealed numerous cohesin and dockerin modules, the basic elements of the cellulosome, and manual sequencing of partially sequenced genomic segments revealed two large tandem scaffoldin-coding genes that form part of a gene cluster. Representative R. champanellensis dockerins were tested against putative cohesins, and the results revealed three different cohesin-dockerin binding profiles which implied two major types of cellulosome architectures: (i) an intricate cell-bound system and (ii) a simplistic cell-free system composed of a single cohesin-containing scaffoldin. The cell-bound system can adopt various enzymatic architectures, ranging from a single enzyme to a large enzymatic complex comprising up to 11 enzymes. The variety of cellulosomal components together with adaptor proteins may infer a very tight regulation of its components. The cellulosome system of the human gut bacterium R. champanellensis closely resembles that of the bovine rumen bacterium Ruminococcus flavefaciens. The two species contain orthologous gene clusters comprising fundamental components of cellulosome architecture. Since R. champanellensis is the only human colonic bacterium known to degrade crystalline cellulose, it may thus represent a keystone species in the human gut.
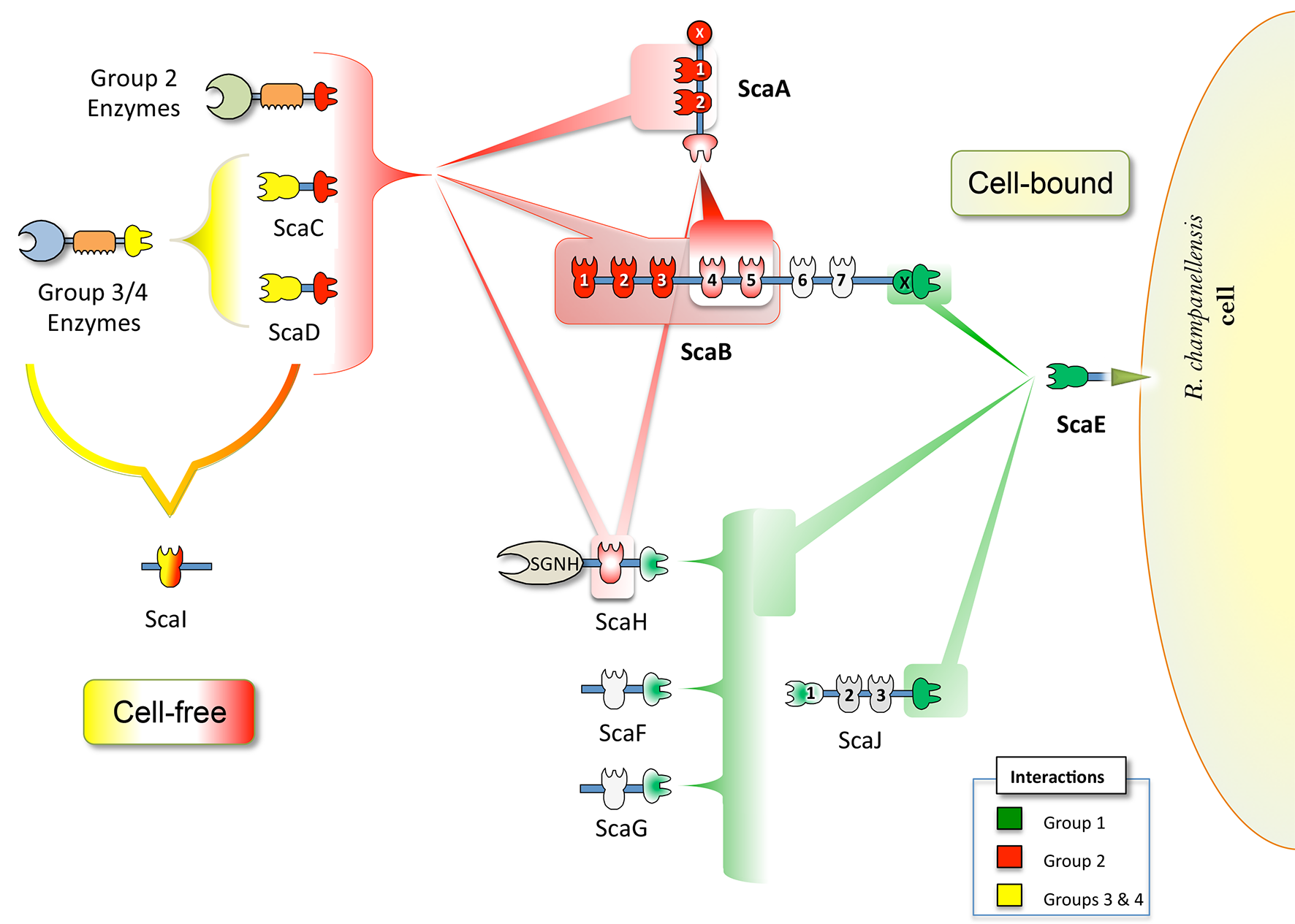
Figure 1. Proposed cell-bound and cell-free cellulosome complexes in R. champanellensis. Different types of cohesin-dockerin interactions are color-coded. The binding specificities of cohesin modules of ScaB6/7, ScaJ2/3, ScaF and ScaG (shown in light gray) are yet to be determined. SGNH stands for a lipase/esterase.
Resistant starch degrader
Ruminococcus bromii is a dominant member of the human gut microbiota that plays a key role in releasing energy from dietary starches that escape digestion by host enzymes via its exceptional activity against particulate “resistant” starches. Genomic analysis of R. bromii shows that it is highly specialized, with 15 of its 21 glycoside hydrolases belonging to one family (GH13). We found that amylase activity in R. bromii is expressed constitutively, with the activity seen during growth on fructose as an energy source being similar to that seen with starch as an energy source. Six GH13 amylases that carry signal peptides were detected by proteomic analysis in R. bromii cultures. Four of these enzymes are among 26 R. bromii proteins predicted to carry dockerin modules, with one, Amy4, also carrying a cohesin module. Since cohesin-dockerin interactions are known to mediate the formation of protein complexes in cellulolytic ruminococci, the binding interactions of four cohesins and 11 dockerins from R. bromii were investigated after overexpressing them as recombinant fusion proteins. Dockerins possessed by the enzymes Amy4 and Amy9 are predicted to bind a cohesin present in scaffoldin 2 (Sca2), which resembles the ScaE cell wall- anchoring protein of a cellulolytic relative, R. flavefaciens. Further complexes are predicted between the dockerin-carrying amylases Amy4, Amy9, Amy10, and Amy12 and two other cohesin-carrying proteins, while Amy4 has the ability to autoaggregate, as its dockerin can recognize its own cohesin. This organization of starch-degrading enzymes is unprecedented and provides the first example of cohesin-dockerin interactions being involved in an amylolytic system, which we refer to as an “amylosome.”
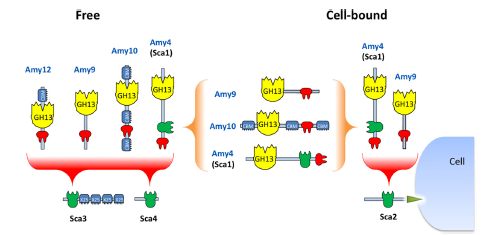
Figure 2. Potential dockerin-mediated interactions of R. bromii amylases with cohesin-carrying proteins and with each other. On the basis of observed interactions with recombinant cohesins and dockerins (Table 4 and Fig. 5), we can predict that the Amy4 and Amy9 enzymes are likely to bind to the cell surface via the Sca2 scaffoldin protein. Further complexes are likely to form between the Amy4, Amy9, and Amy10 proteins and between Amy4, Amy9, Amy10, and Amy12 and the Sca3 and Sca4 proteins. Binding of the enzymes to Sca3 would presumably confer starch-binding features to the resultant complex. Binding of dockerin-bearing amylases Amy9 and Amy10 to the cohesin of Amy4 would result in the formation of multienzyme complexes that can be part of a cell-bound or cell-free system. Amy4 also has the potential to self-aggregate through interactions between its own cohesin and dockerin.
Future research directions:
- How widely distributed are these organisms in the human gut microbiome?
- How significant is cellulose/starch degradation in the human gut?
- Does the degradation of these substrates provide a significant source of energy to the human host or is this degradation an upstream process, e.g., to prevent damage to the soft tissue of our digestive tract from the abrasive cellulose fibers that we ingest in our fruit and vegetables?
More on that subject:
http://wis-wander.weizmann.ac.il/life-sciences/me-and-my-cellulosome
- Michel G (2015) Ruminococcal cellulosomes: molecular Lego to deconstruct microcrystalline cellulose in human gut. Environ Microbiol 17:3113–3115. doi: 10.1111/1462-2920.12920
- Cann I, Bernardi RC, Mackie RI (2016) Cellulose degradation in the human gut: Ruminococcus champanellensis expands the cellulosome paradigm. Environ Microbiol n/a–n/a. doi: 10.1111/1462-2920.13152
- Moraïs S, David Y Ben, Bensoussan L, et al. (2016) Enzymatic profiling of cellulosomal enzymes from the human gut bacterium, Ruminococcus champanellensis , reveals a fine-tuned system for cohesin-dockerin recognition. Environ Microbiol 18:542-56. doi: 10.1111/1462-2920.13047
- Ben David Y, Dassa B, Borovok I, et al. (2015) Ruminococcal cellulosome systems from rumen to human. Environ Microbiol 17:n/a–n/a. doi: 10.1111/1462-2920.12868
- Ze X, David B, Laverde-gomez JA, et al. (2015) Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic firmicutes bacterium Ruminococcus bromii. MBio 6:1–11. doi: 10.1128/mBio.01058-15.Editor



