The chemical and structural intricacy of plant cell wall polysaccharides is matched by the diversity and complexity of the enzymes that degrade them. Cellulases and hemicellulases are family members of the broad group of glycoside hydrolases, which catalyze the hydrolysis of oligosaccharides and polysaccharides in general. These enzymes are composed of a series of separate modules. This fact explains the very large size of some of these enzymes and gives us some insight into their complex mode of action. Each module or domain comprises a consecutive portion of the polypeptide chain and forms an independently folding, structurally and functionally distinct unit. Each enzyme contains at least one catalytic module, which catalyzes the actual hydrolysis of the glycosidic bond and provides the basis for classification of the simple enzymes. Other accessory or "helper" domains assist or modify the primary hydrolytic action of the enzyme, thus modulating the overall properties of the enzyme. Some of the different themes illustrating the modular compositions of the cellulases and related enzymes are illustrated in Figure 1.
The major difference between free enzymes and cellulosomal enzymes is that the free enzymes usually contain a CBM for guiding the catalytic domain to the substrate, whereas the cellulosomal enzymes carry a dockerin domain that incorporates the enzyme into the cellulosome complex. Otherwise, both the free and cellulosomal enzymes contain very similar types of catalytic domains. The cellulosomal enzymes rely on the Family-3a CBM of the scaffoldin subunit for collective binding to crystalline cellulose.
Today, well over 100 glycoside hydrolase families of and over 50 CBM families have been described. For more details, see our favorite Website on Carbohydrate-Active enZymes – CAZy (http://www.cazy.org/).
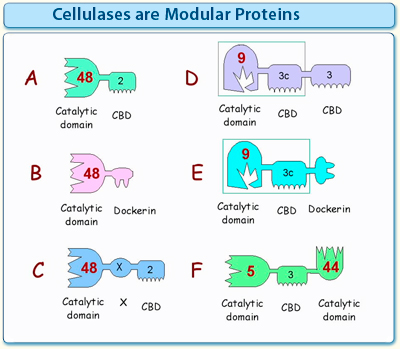
Figure 1. Simplistic scheme illustrating the diversity of the modular architecture of cellulases and other glycoside hydrolases. The different modules are grouped into families according to conserved sequences.
- One of the most common types of “free” (non-cellulosomal) cellulases consists of a catalytic module, flanked by a CBM at its N- or C-terminus.
- Cellulosomal enzymes are characterized by a dockerin module attached to a catalytic domain.
- Many cellulases contain "X modules", i.e., modules of unknown (as yet undefined) function.
- Some enzymes have more than one CBM. Often, one CBM serves to target the cellulase to the flat surface of the insoluble substrate, whereas the other acts in concert with the catalytic module by binding transiently to a single cellulose or hemicellulose chain.
- Some cellulosomal cellulases have a CBM together with a dockerin in the same polypeptide chain.
- Some cellulases have more than one type of catalytic module in the same polypeptide chain, together with one or more CBMs and/or a dockerin.
The cellulases include the large number of endo- and exo-glucanases which hydrolyze -1,4-glucosidic bonds of the cellulose chains. Thus, in principle, the degradation of cellulose requires the cleavage of a single type of bond. Nevertheless, in practice, we find that cellulolytic microorganisms produce a variety of complementary cellulases of different specificities from many different families. Cellulases that degrade the cellulose chain can be either "endo-acting", or "exo-acting". The distinction between endo- and exo-acting enzymes is also reflected by the architecture of the respective class of active site, even within the same family of enzymes (Fig. 2). The endoglucanases, for example, are commonly characterized by a groove or cleft, into which any part of a linear cellulose chain can fit. On the other hand, the exoglucanases bear tunnel-like active sites, which can only accept a substrate chain via its terminus. The exo-acting enzyme apparently threads the cellulose chain through the tunnel, wherein successive units (e.g., cellobiose) would be cleaved in a sequential, “processive” manner. The different modes of action have been suggested to account for the observed synergy among the different types of cellulases. For example, an endo-acting enzyme can produce new chain ends in the internal portion of the cellulose chain, and the two newly exposed chains would then be available for action of exo-acting enzymes. In addition, two different types of exo-glucanases may exhibit different specificities by acting on a cellulose chain from opposite ends (i.e., the reducing versus the nonreducing end).
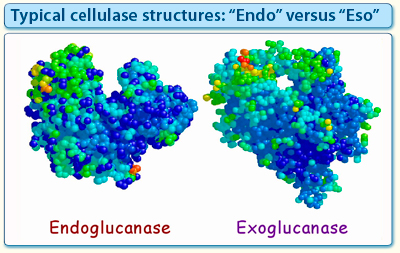
Figure 2. Structures of a typical Family-6 endoglucanase and exoglucanase. Despite the sequence similarity of both enzymes, their respective active-site architecture is different. The endoglucanase exhibits a deep cleft to accommodate the cellulose chain at any point along its length, whereas the active site of the exoglucanase bears an extended loop that forms a tunnel, through which one of the termini of a cellulose chain can be threaded.
In addition to endo- and exo-glucanases, the cellulases include β-glucosidases, which hydrolyze cellobiose — the major endproduct of cellulase digestion — to generate two molecules of glucose. Often, the β-glucosidases are associated with the microbial cell surface and hydrolyze cellobiose to glucose as the cellodextrins are transported into the cell.
Hemicellulases
In contrast to cellulose degradation, the degradation of the hemicelluloses imposes a somewhat different challenge, since this group of polysaccharides includes widely different types of sugars or non-sugar constituents with different types of bonds. Hemicellulases can be divided into two main types, those that cleave the mainchain backbone, i.e. xylanases or mannanases, and those that degrade sidechain substituents or short end products, such as arabinofuranosidase, glucuronidase and xylosidase. Like the cellulases, hemicellulases can be of the endo or exo types.
Some of our research projects on cellulose-degrading enzymes
Studies on the mechanism of action of a processive endoglucanase. Family-9 cellulases are a very broad and interesting group of enzymes, which allow us to examine the transition of a simple endoglucanase to the “processive” mode of action, whereby the substrate is degraded in a systematic manner. The different modular themes of the family-9 cellulases are given in Fig. 3. In the past, we have examined the mechanism of action of the non-cellulosomal C. thermocellum enzyme, Cel9I. In recent work, we have successfully combined separate catalytic and ancillary CBM3c modules, resulting in a non-covalent association of the two. We are currently examining the biochemical and catalytic properties of the associated components. In addition, we have obtained diffracting crystals, and we hope to determine the first 3D structure of functional non-covalently coupled modules. In doing so, we expect to gain insight into the molecular features responsible for altering the mode of cellulolytic activity in this important cellulase family. See the following articles for more information:
Gilad, R., Rabinovich, L., Yaron, S., Bayer, E. A., Lamed, R., Gilbert, H. J., and Shoham, Y. (2003) CelI, a non-cellulosomal family-9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185, 391-398.
Jindou, S., Xu, Q., Kenig, R., Shoham, Y., Bayer, E. A., and Lamed, R. (2006) Novel architectural theme of family-9 glycoside hydrolases identified in cellulosomal enzymes of Acetivibrio cellulolyticus and Clostridium thermocellum. FEMS Microbiol. Lett. 254, 308-316.
Jindou, S., Petkun, S., Shimon, L., Bayer, E. A., Lamed, R., and Frolow, F. (2007) Crystallization and preliminary diffraction studies of CBM3b of cellobiohydrolase 9A from Clostridium thermocellum. Acta Crystallogr. F63, 1044–1047.
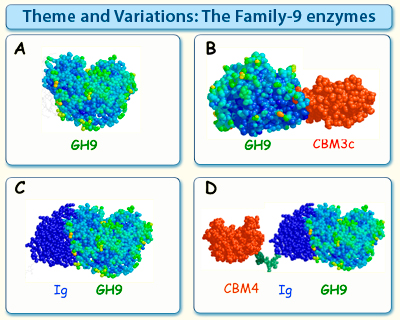
Figure 3. The family-9 enzymes can be subdivided into several thematic types, based on their modular structures.
Theme A enzymes contain the catalytic module, usually attached to either a dockerin or cellulose-specific CBM (not shown), without additional ancillary modules.
Theme B enzymes contain a family-3c CBM fused to the family-9 catalytic module at its C-terminus, separated therefrom by a characteristic linker segment. In certain cases, the CBM aligns in such a manner that the endoglucanase character of the catalytic module is altered to act in a processive manner.
Theme C enzymes contain an Ig domain fused to the catalytic module at its N-terminus, presumably providing extra stability to the endoglucanase.
Theme D enzymes resemble the Theme C enzymes with an added family-4 CBM on the N-terminus of the Ig domain. The exact structure of this type of enzyme with its modular parts has yet to be elucidated.
Enzymatic profiling of cellulosomal enzymes
In a recent [1] study, we cloned, expressed and characterized all of the glycoside hydrolases that contain a dockerin module in a single bacterium. The enzymes of R. champanellensis were expressed in small scale and tested against a variety of polysaccharides substrates. Among the 25 enzymes: 10 cellulases, 4 xylanases, 3 mannanases, 2 xyloglucanases, 2 arabinofuranosidases, 2 arabinanases and one β- glucanase were revealed and assessed for their comparative enzymatic activity on their respective substrates. The binding specificities of the dockerin modules were also examined by ELISA, and 80 positives out of 525 possible interactions were detected. Our analysis reveals a fine-tuned system for cohesin-dockerin specificity and the importance of diversity among the cohesin-dockerin sequences. Our results imply that cohesin-dockerin pairs are not necessarily assembled at random among the same specificity types, as generally believed for other cellulosome-producing bacteria, but reveal a more organized cellulosome architecture.
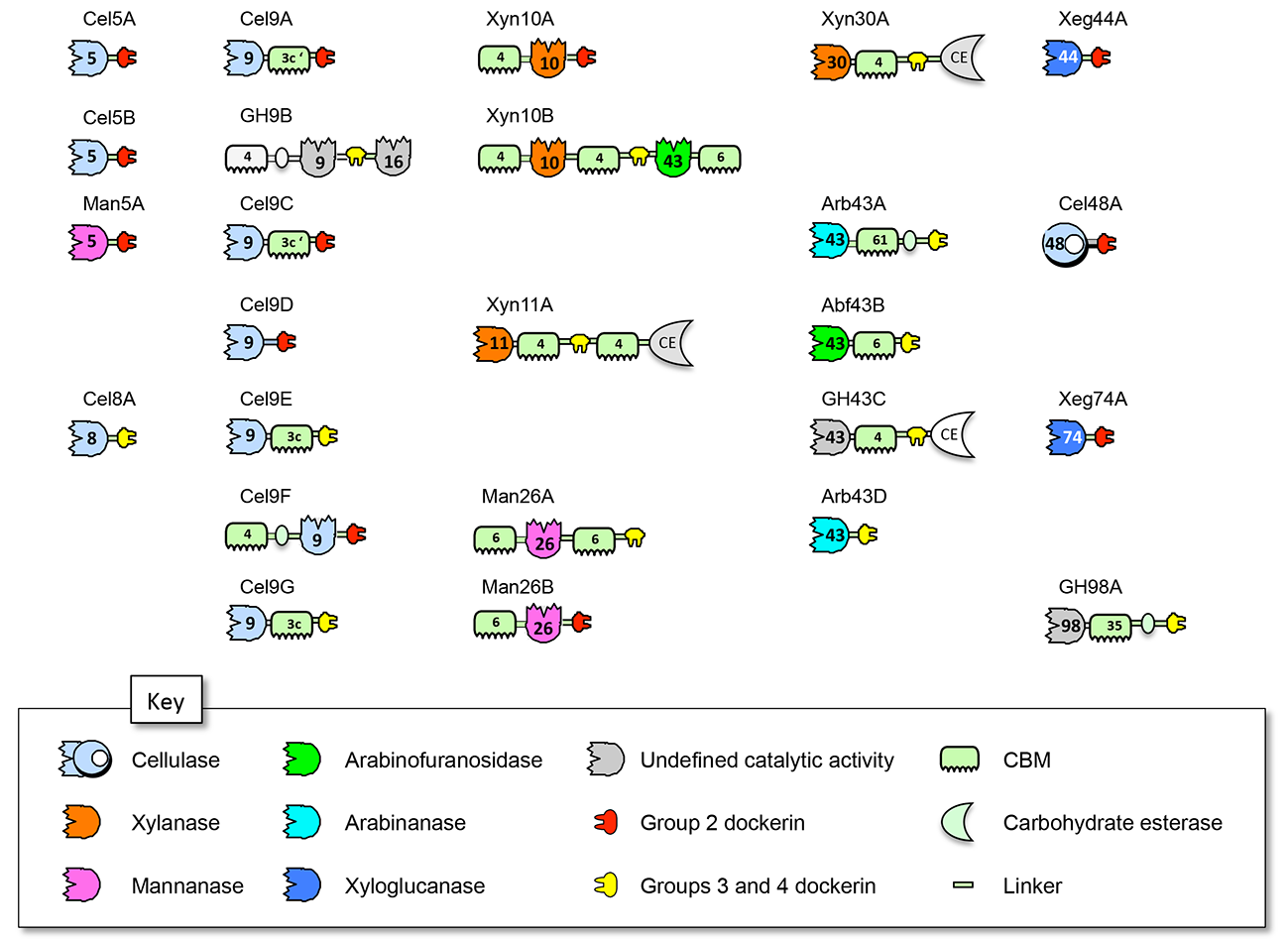
Figure 4: Schematic representation and proposed nomenclature of the dockerin- containing glycoside hydrolases from R. champanellensis. Enzyme activity and dockerin-specificity are color-coded. GH family or CBM numbers are indicated.
Moraïs S, David Y Ben, Bensoussan L, et al. (2016) Enzymatic profiling of cellulosomal enzymes from the human gut bacterium, Ruminococcus champanellensis, reveals a fine-tuned system for cohesin-dockerin recognition. Environ Microbiol 18:542-56. doi: 10.1111/1462-2920.13047



