Single Molecule Spectroscopy on Immobilized Molecules
To extend dramatically the measurement time of individual molecules, while minimally affecting their conformational dynamics, we encapsulate them within phospholipid vesicles. These vesicles are in turn tethered to the surface using streptavidin-biotin chemistry.


Protein Folding and Misfolding
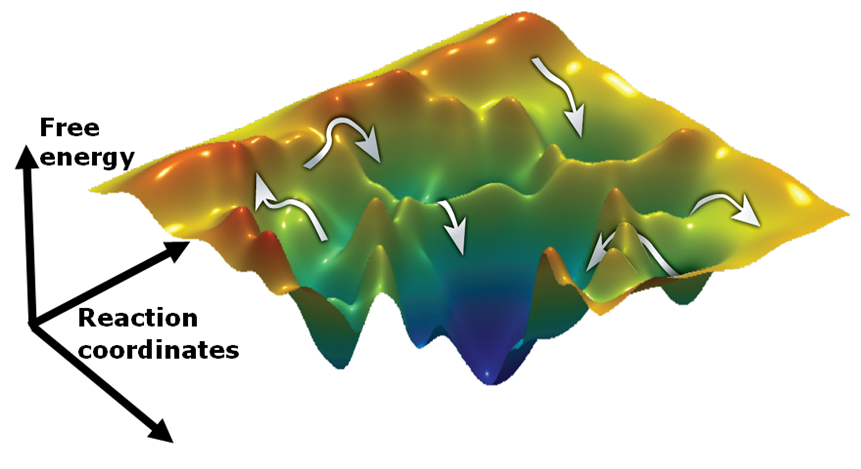
As a step towards understanding the role and function of such misfolded proteins, the process of protein folding and the mechanisms underlying it must be described. Protein folding is considered a complex issue and one way to address it is by studying the simple case of small proteins (e.g Protein L), which possess a simple energy landscape. In the same manner we can follow much more complicated folding processes by studying large multidomain proteins such as adenylate kinase (AK).
Conformational Dynamics
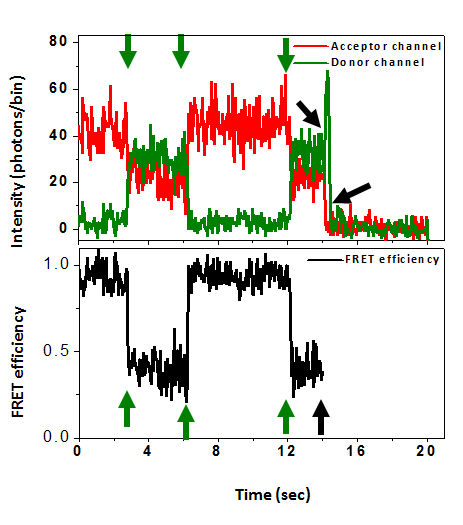
Protein dynamics often involve the relative motion of two structural units (domains). To understand principles underlay such motions, we study the hinge bending motion of AK during its working cycle. Also, we seek to understand the impact of clinically-relevant mutations on the function and dynamics of human adenylate kinase 1.
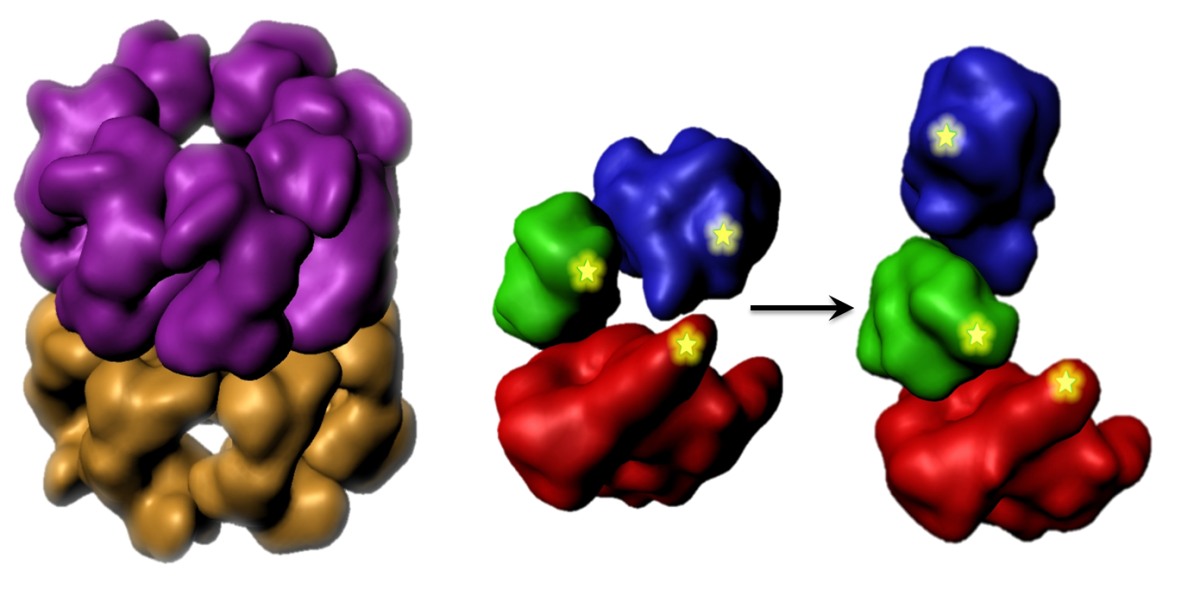
Large protein machines coordinate their subunit motions in order to function properly. We seek to measure the temporal order and extent of domain and subunit motions within selected machines such as the molecular chaperone GroEL.