Evolution of the noncoding genome
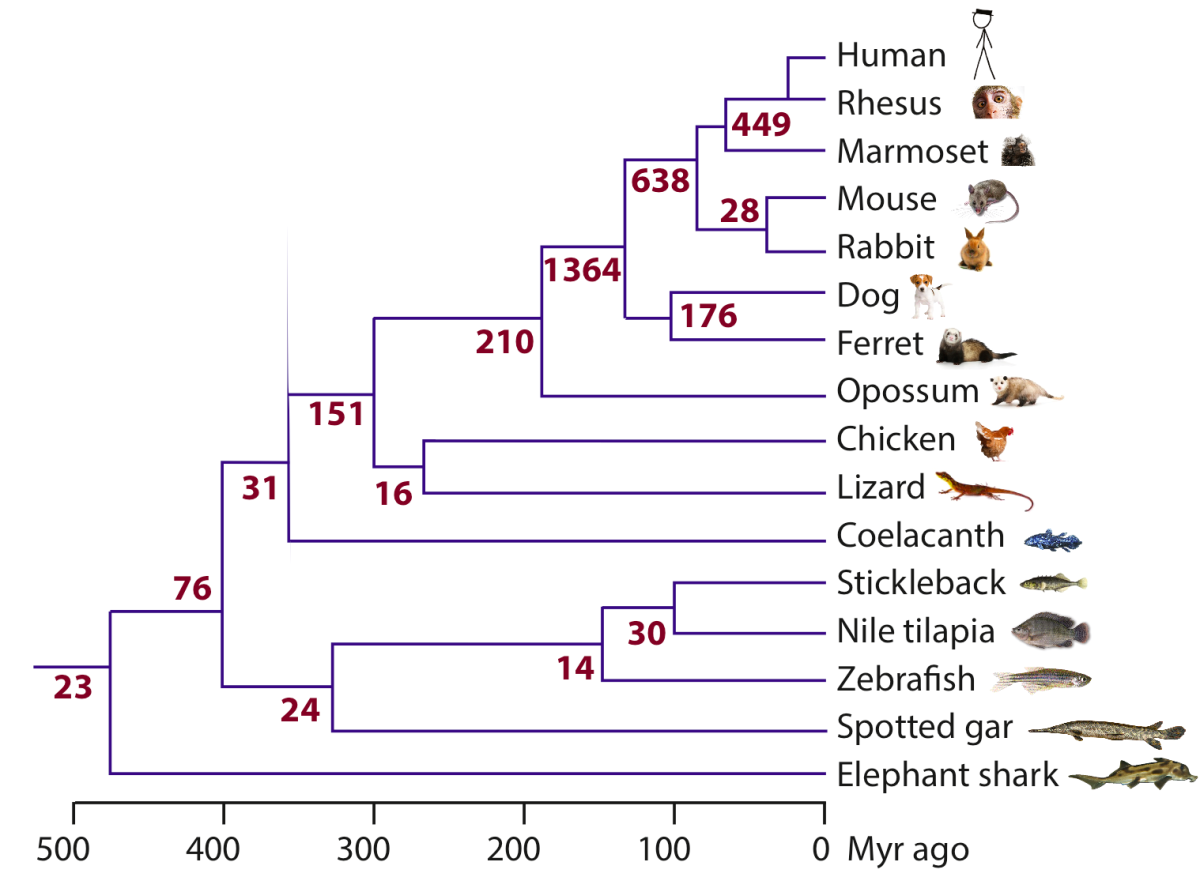
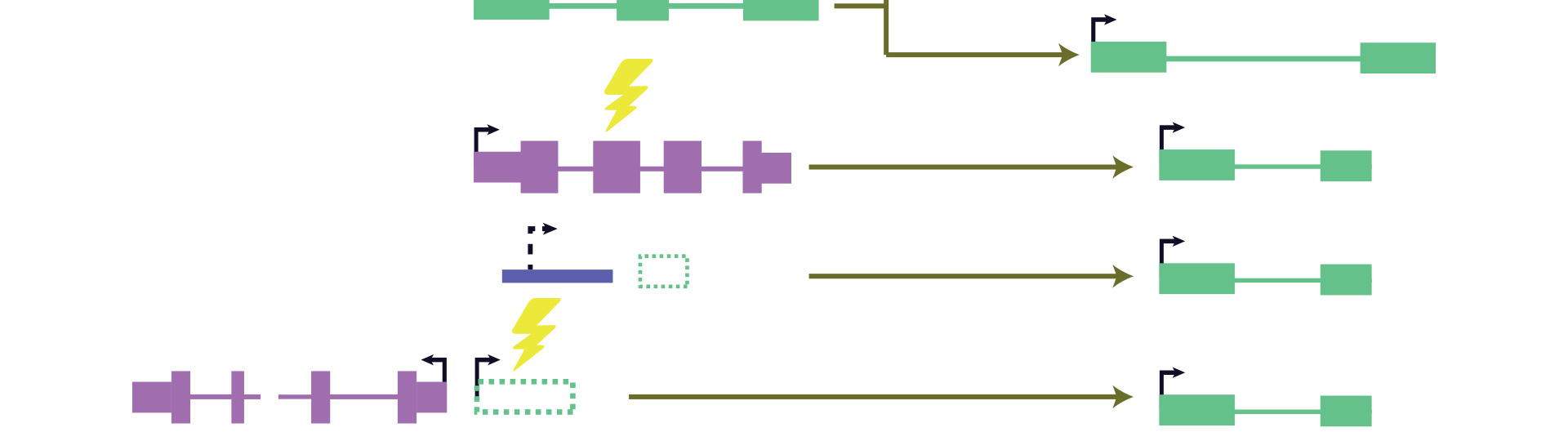
Comparative analysis has played instrumental roles in allowing us to understand the rules underlying protein and microRNA function, but it has been difficult to apply the existing methods to study lncRNAs because of scarcity of lncRNA annotations and rapid lncRNA evolution (see review). To build a catalog of lncRNAs in vertebrates, we collected RNA-seq and 3P-seq data and developed robust methods for lncRNA identification in 17 species (Hezroni et al., 2015). We also developed methods for identifying lncRNA homologs based on subtle sequence homology and conservation of neighboring genes and genomic context. Using these catalogs, we showed that rapid turnover of most lncRNA genes is accompanied by positional conservation in some lncRNAs and retention of short islands of sequence conservation in others. We then focused on the evolutionary origins of those lncRNAs that are found only in mammals and traced ~5% of them to protein-coding genes that had lost their coding potential before the rise of mammals (Hezroni et al., 2017). These pseudogene-derived lncRNAs are associated with features that set them apart from other lncRNAs, such as broader expression domains. We are developing methods for mining the sequences of those lncRNAs that are conserved in sequence or in position and for identifying conserved combinations of short sequence and/or structure elements that are undetectable using existing approaches designed for protein-coding genes (Ross, Genome Biology 2021 and ongoing).