Cancer Omics is largely dominated by genomic approaches. However, multiple studies have reported an average correlation between protein and RNA expression of ~0.5 across patient samples. In our previous proteogenomic studies we found that unsupervised clustering of clinical breast tumors based on proteomics data a novel breast cancer subtypes that is not found in RNA datasets (Yanovich et al, Cancer Research 2018); and analysis of melanoma samples identified a unique association between mitochondrial metabolism and cancer immunogenicity (Harel et al, Cell 2019). Furthermore, a comparison between proteomic internal heterogeneity and genomic heterogeneity unravelled markedly reduced heterogeneity at the proteomic level, thereby overcoming stochastic, non-functional genomic changes. Overall, these studies show the important contribution of the proteomic layer to our understanding of cancer dynamics, and the power of using proteomics as the basis for future drug development.
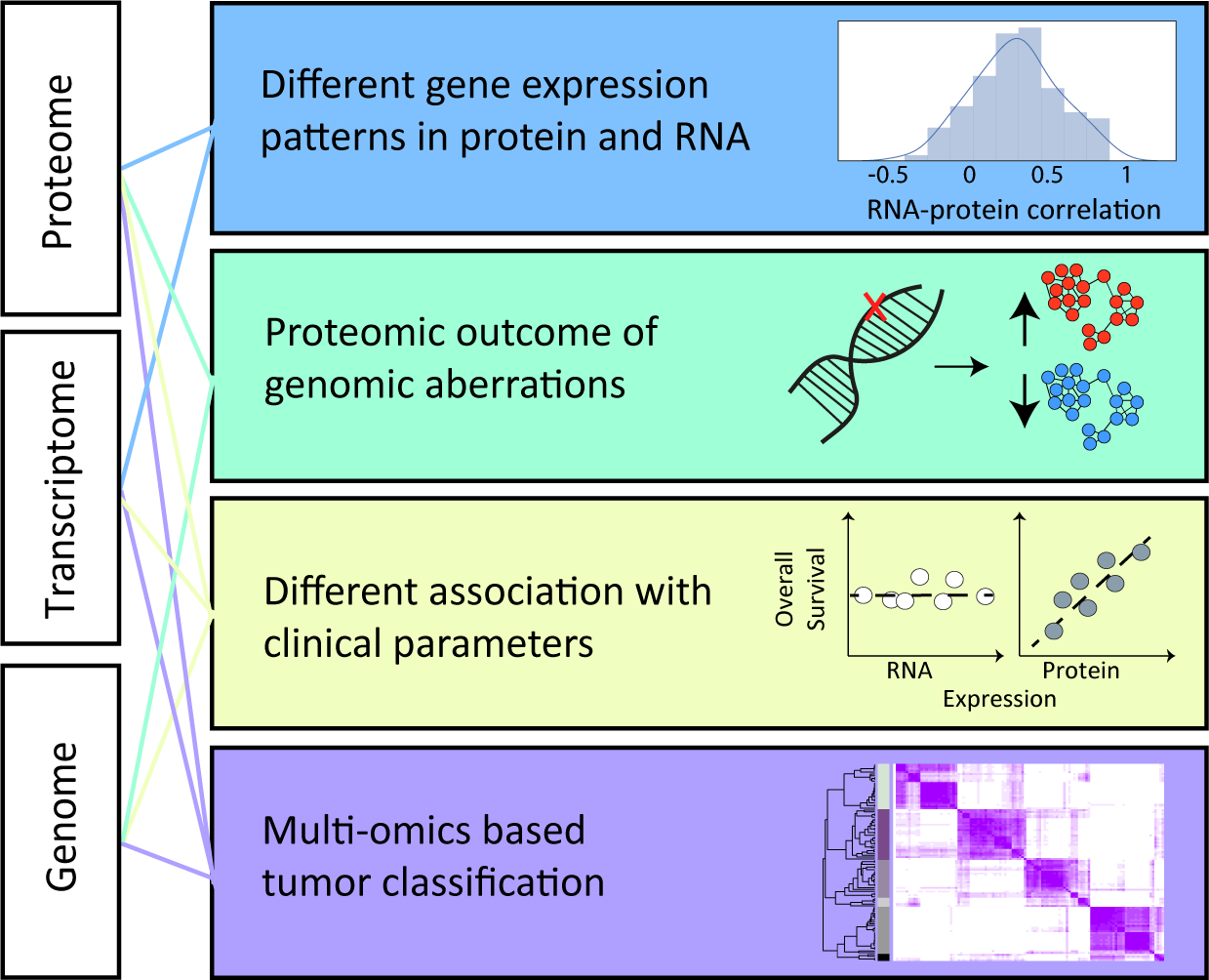
Proteogenomic analyses combine genome-scale proteomics with genomics and/or transcriptomics. Comparison among layers can unravel distinct expression levels and mechanisms of expression regulation, which might reveal the output of genomic aberrations in cancer. Proteogenomic analyses can also associate each layer with the patient clinical parameters and identify novel cancer subtypes.