Analyzing single cells with mass spectrometry based proteomics presents an enormous technological challenge. Reaching deep proteomic coverage from sub-nanogram protein amounts from single cells requires development of novel mass spectrometric approaches, sample preparation techniques and computational analyses. We develop workflows for deep proteomics, and apply these approaches to tumor samples from cancer mouse models and human tumors. Analyses of thousands of cells per tumor are then able to unravel the internal cancer heterogeneity and the interactions between the cancer cells and the microenvironment. Single cell analyses can then provide the basis for identification of drug resistance mechanisms and development of novel therapeutic approaches.
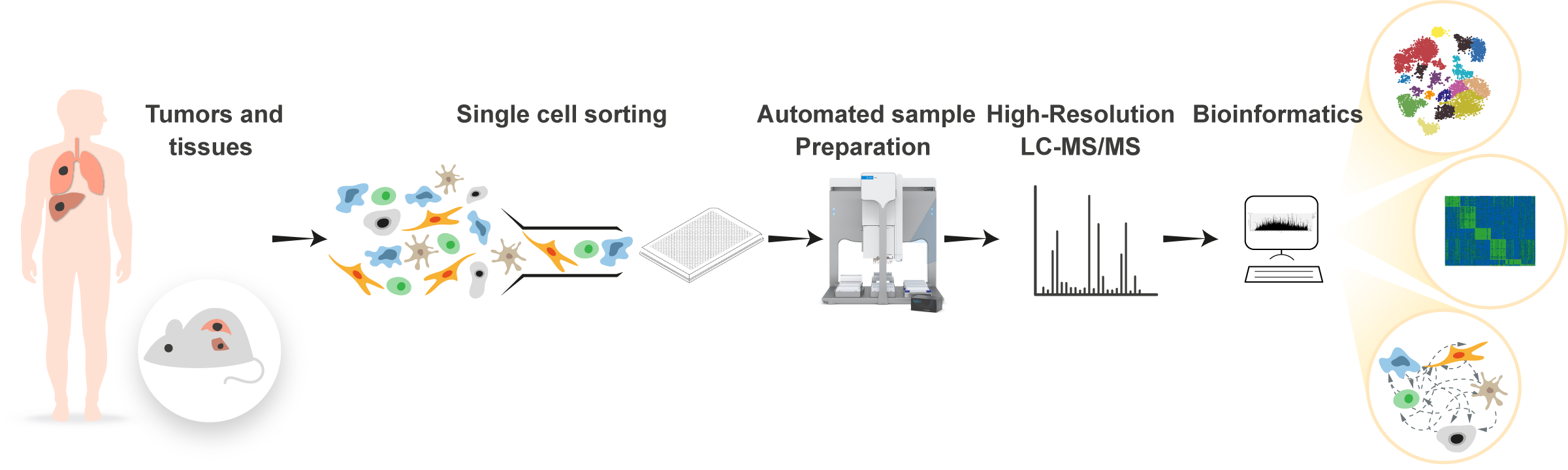
Automated single cell proteomics workflow.