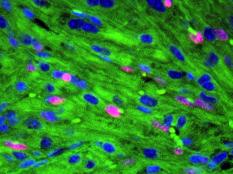
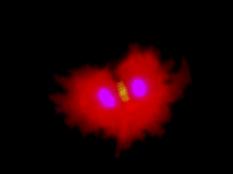
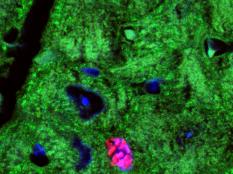
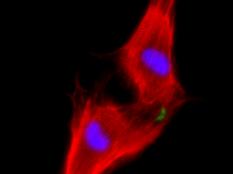
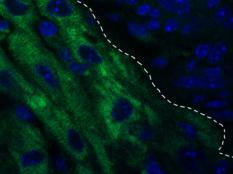
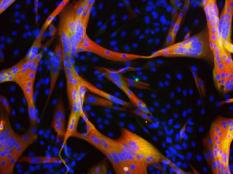
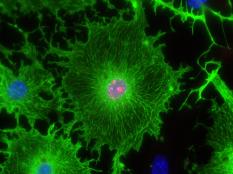
Our lab forms a bridge between developmental biology and regenerative medicine aiming to develop novel approaches for mammalian heart regeneration
Tzahor Lab
Home
About us
Selected Publications
Research Areas
Research
Cardiomyocyte cell-cycle control and dedifferentiation
Ischemic heart disease is the most common cardiac ailment, and the leading cause of death in the Western world today. We combine novel approaches to study cardiomyocyte (CM) renewal by promoting CM cell division and dedifferentiation, as a potential therapeutic strategy to cure the injured heart. We focus on various signaling strategies (e.g., NRG/ERBB signaling pathway), combined with attention to the microenvironment. We focus on the biophysical properties (e.g., matrix rigidity) of the heart microenvironment as well on biochemical contents of the ECM and ECM- associated molecules. Finally, we employ a novel state-of-the-art high-throughput screening platform to identify molecules that promote CM proliferation and to study their effects on cardiac regeneration in mice. We have identified several novel compounds which significantly increase the proliferation of adult CMs. Understanding how mature CMs can disassemble their sarcomeric architecture and re-enter the cell cycle, combined with the development of novel procedures that can facilitate CM dedifferentiation and proliferation, are major challenges facing current biomedical research. Our research into myogenesis and cardiogenesis in chick embryos demonstrates that the FGF-ERK signaling pathway plays an inhibitory role in myogenic differentiation. We now seek to understand the molecular and cellular underpinnings of this phenomenon using adult muscle stem cells.
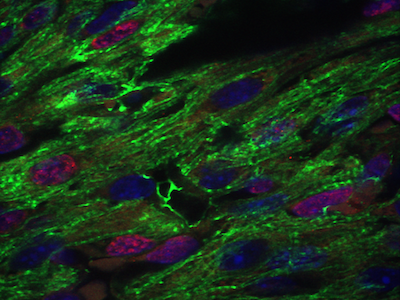
Cardiac and craniofacial development
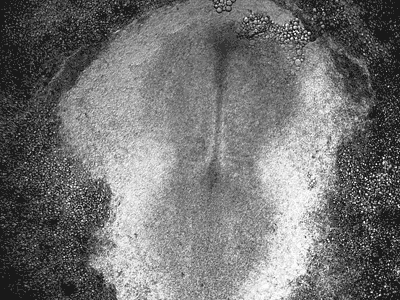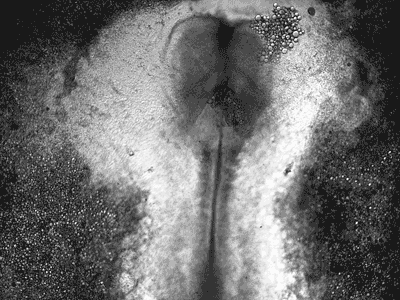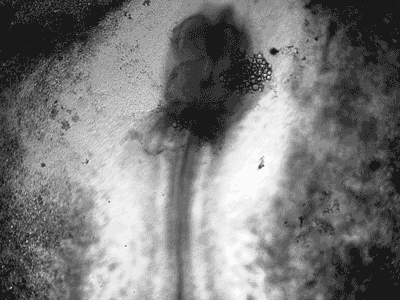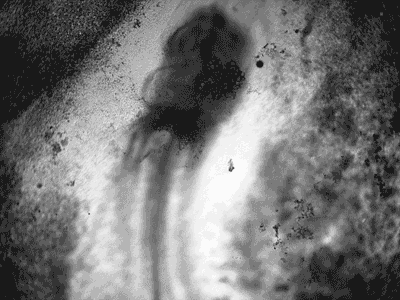
Manipulating tumorigenic processes to stimulate heart regeneration
Perhaps one of the most surprising and interesting aspects of the adult heart is the fact that cardiac tumors rarely develop. This implies that some factor/s within the heart, which we are keen to identify, resists tumorigenesis. Inducing oncogenic transformation of cardiac cells may help to understand the heart resistance to cancer. Unleashing cell cycle control is a hallmark of cancer; on the other hand, these same mechanisms could be very useful in promoting heart regeneration. Hence, another angle of our research focus on the pros and cons of cell-cycle re-entry, finding just the right balance between these two processes (regeneration and tumorigenesis).
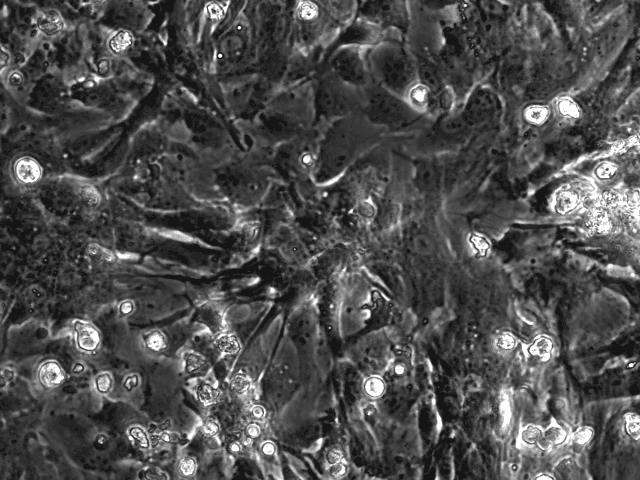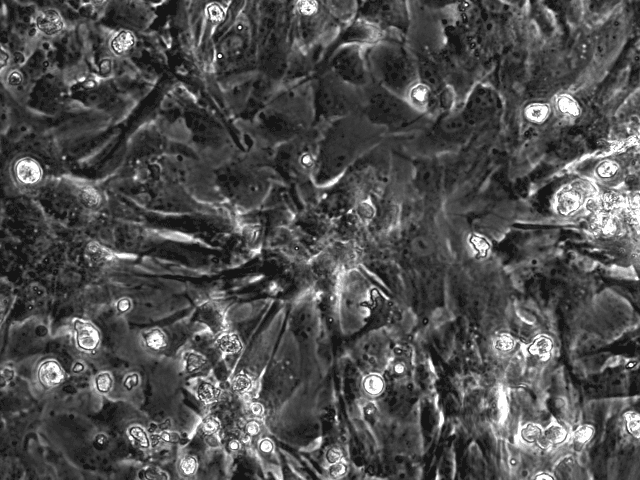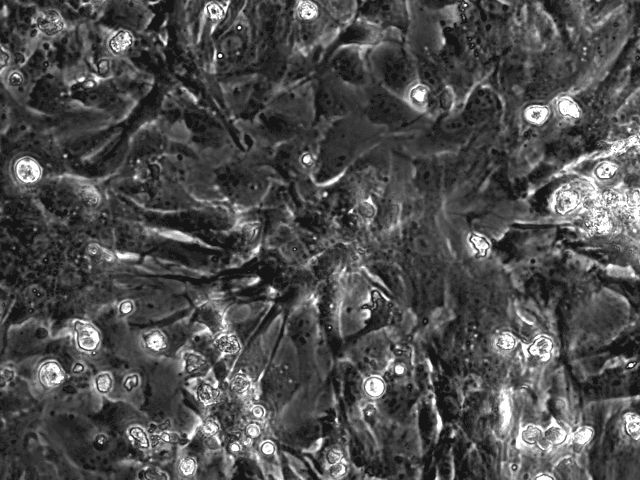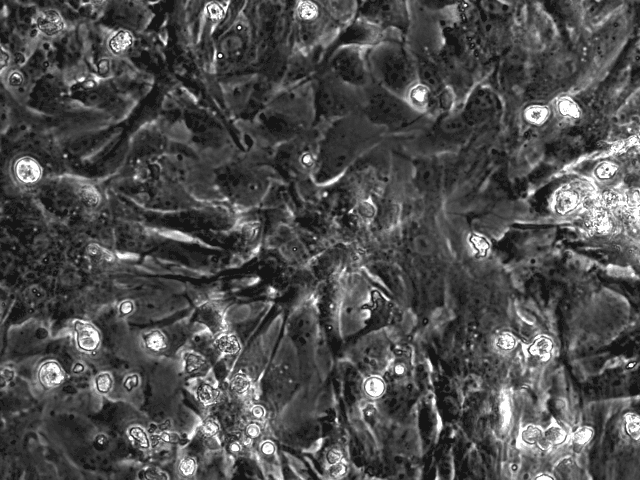
Publications
-
(2024). How Can Young Extracellular Matrix Promote Cardiac Regeneration? Versi-Can!. Circulation. 2024 Mar , 149 (13):1016-1018.
-
(2023). An enhancer-based gene-therapy strategy for spatiotemporal control of cargoes during tissue repair. Cell Stem Cell. 2023 Jan , 30 (1):96-111.e6.
-
(2023). Interplay between calcium and sarcomeres directs cardiomyocyte maturation during regeneration. Science (New York, N.Y.). 2023 May , 380 (6646):758-764.
-
(2023). Redifferentiated cardiomyocytes retain residual dedifferentiation signatures and are protected against ischemic injury. Nature Cardiovascular Research. 2023 Apr , 2 (4):383-398.
-
(2023). The tumor microenvironment shows a hierarchy of cell-cell interactions dominated by fibroblasts. Nature Communications. 2023 Sept , 14 (1).
-
(2022). How many fish make a mouse?. Nature Cardiovascular Research. 2022 Jan , 1 :2-3.
-
(2022). A coalition to heal—the impact of the cardiac microenvironment. Science. 2022 Sept , 377 (6610).
-
(2022). Glucocorticoid receptor antagonization propels endogenous cardiomyocyte proliferation and cardiac regeneration. Nature Cardiovascular Research. 2022 Jul , 1 (7):617-633.
-
(2021). ERK1/2 inhibition promotes robust myotube growth via CaMKII activation resulting in myoblast-to-myotube fusion. Developmental Cell. 2021 Dec , 56 (24):3349-3363.e6.
-
(2021). Myocardial infarction techniques in adult mice. . 2021, :3-21.
-
(2021). The extracellular matrix protein agrin is essential for epicardial epithelial-to-mesenchymal transition during heart development. Development (Cambridge). 2021 May , 148 (9).
-
(2021). Small-molecule inhibition of Lats kinases may promote Yap-dependent proliferation in postmitotic mammalian tissues. Nature Communications. 2021 May , 12 (1).
-
(2020). In remembrance of David Yaffe. Skeletal Muscle. 2020 Oct , 10 (1).
-
(2020). ERBB2 drives YAP activation and EMT-like processes during cardiac regeneration. Nature Cell Biology. 2020 Nov , 22 (11):1346-1356.
-
(2020). Agrin promotes coordinated therapeutic processes leading to improved cardiac repair in pigs. Circulation. 2020 Sept , 142 (9):868-881.
-
(2019). MBP-FGF2-Immobilized Matrix Maintains Self-Renewal and Myogenic Differentiation Potential of Skeletal Muscle Stem Cells. International Journal of Stem Cells. 2019 Jul , 12 (2):360-366.
-
(2019). Transient p53-Mediated Regenerative Senescence in the Injured Heart. Circulation. 2019 May , 139 (21):2491-2494.
-
(2019). Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. eLife. 2019 Dec , 8 .
-
(2019). The small molecule Chicago Sky Blue promotes heart repair following myocardial infarction in mice. JCI insight. 2019 Nov , 4 (22).
-
(2019). Metabolic modulation regulates cardiac wall morphogenesis in zebrafish. eLife. 2019 Dec , 8 .
-
(2019). Vitamin D Stimulates Cardiomyocyte Proliferation and Controls Organ Size and Regeneration in Zebrafish. Developmental Cell. 2019 Mar , 48 (6):853-863.e5.
-
(2018). Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation. Nature. 2018 May , 557 (7705):439-+.
-
(2017). Cardiac regeneration strategies: Staying young at heart.. SCIENCE. 2017, 356 (6342):1035-1039.
-
(2017). Nkx2.5 marks angioblasts that contribute to hemogenic endothelium of the endocardium and dorsal aorta. eLife. 2017 May , .
-
(2017). The cancer paradigms of mammalian regeneration: Can mammals regenerate as amphibians?. Carcinogenesis. 2017 Apr , 38 (4):359-366.
-
(2017). Cardiac regeneration strategies: Staying young at heart. Science. 2017 Jun , 356 (6342):1035-1039.
-
(2017). The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017 Jul , 547 (7662):179-184.
-
(2016). Coronary vasculature patterning requires a novel endothelial ErbB2 holoreceptor. Nature Communications. 2016 Jun , 7 .
-
(2016). Macrophage precursor cells from the left atrial appendage of the heart spontaneously reprogram into a C-kit +/CD45 - Stem cell-like phenotype. International Journal of Cardiology. 2016 Apr , 209 :296-306.
-
(2016). Loss of Muscle MTCH2 Increases Whole-Body Energy Utilization and Protects from Diet-Induced Obesity. Cell Reports. 2016 Feb , 14 (7):1602-1610.
-
(2015). A new heart for a new head in vertebrate cardiopharyngeal evolution. Nature. 2015 Apr , 520 (7548):466-473.
-
(2015). Head Muscle Development. . 2015, :123-142.
-
(2015). ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nature Cell Biology. 2015 May , 17 (5):627-638.
-
(2015). Reduced matrix rigidity promotes neonatal cardiomyocyte dedifferentiation, proliferation and clonal expansion.. eLife. 2015, 4 .
-
(2015). The key roles of ERBB2 in cardiac regeneration. Cell Cycle. 2015 Jan , 14 (15):2383-2384.
-
(2015). Craniofacial Muscle Development. . 2015, :3-30.
-
(2014). Endothelial cells regulate neural crest and second heart field morphogenesis. Biology Open. 2014 Aug , 3 (8):679-688.
-
(2014). Nuclear to cytoplasmic shuttling of ERK promotes differentiation of muscle stem/progenitor cells. Development (Cambridge). 2014 Jul , 141 (13):2611-2620.
-
(2013). Chronic Akt1 deficiency attenuates adverse remodeling and enhances angiogenesis after myocardial infarction. Circulation-Cardiovascular Imaging. 2013 Nov , 6 (6):992-1000.
-
(2012). Pharyngeal mesoderm regulatory network controls cardiac and head muscle morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012 Nov , 109 (46):18839-18844.
-
(2012). The actin regulator N-WASp is required for muscle-cell fusion in mice. Proceedings of the National Academy of Sciences of the United States of America. 2012 Jul , 109 (28):11211-11216.
-
(2011). Pharyngeal mesoderm development during embryogenesis: Implications for both heart and head myogenesis. Cardiovascular Research. 2011 Jul , 91 (2):196-202.
-
(2011). p53 and epithelial-mesenchymal transition: A linking thread between embryogenesis and cancer. Cell Cycle. 2011 Sept , 10 (18):3036-3037.
-
(2011). P53 coordinates cranial neural crest cell growth and epithelial-mesenchymal transition/delamination processes. Development. 2011 May , 138 (9):1827-1838.
-
(2011). The heart endocardium is derived from vascular endothelial progenitors. Development. 2011 Nov , 138 (21):4777-4787.
-
(2010). The occipital lateral plate mesoderm is a novel source for vertebrate neck musculature. Development. 2010 Sept , 137 (17):2961-2971.
-
(2010). BMP-mediated inhibition of FGF signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development. 2010 Sept , 137 (18):2989-3000.
-
(2009). Hypoxia-Inducible Factor-1 alpha and-2 alpha Additively Promote Endothelial Vasculogenic Properties. Journal of Vascular Research. 2009, 46 (4):299-310.
-
(2009). sFRPs: A declaration of (Wnt) independence. Nature Cell Biology. 2009, 11 (1):13-14.
-
(2009). p53 involvement in early cranial neural crest development in the chick. Developmental Biology. 2009 Jul , 331 (2):430-430.
-
(2009). Heart and craniofacial muscle development: A new developmental theme of distinct myogenic fields. Developmental Biology. 2009 Mar , 327 (2):273-279.
-
(2009). Isl1 is a direct transcriptional target of Forkhead transcription factors in second heart field-derived mesoderm. Developmental Biology. 2009 Oct , 334 (2):513-522.
-
(2009). Distinct Origins and Genetic Programs of Head Muscle Satellite Cells. Developmental Cell. 2009 Jun , 16 (6):822-832.
-
(2008). The contribution of Islet1-expressing splanchnic mesoderm cells to distinct branchiomeric muscles reveals significant heterogeneity in head muscle development. Development. 2008 Feb , 135 (4):647-657.
-
(2008). Reconstruction of Cell Lineage Trees in Mice. PLoS ONE. 2008 Apr , 3 (4).
-
(2008). P53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS ONE. 2008 Nov , 3 (11):1-15.
-
(2008). Constitutive expression of HIF-1α and HIF-2α in bone marrow stromal cells differentially promotes their proangiogenic properties. Stem Cells. 2008 Oct , 26 (10):2634-2643.
-
(2008). Estimating cell depth from somatic mutations. PLoS Computational Biology. 2008 May , 4 (5).
-
(2007). Cranial neural crest cells regulate head muscle patterning and differentiation during vertebrate embryogenesis. Development. 2007 Sept , 134 (17):3065-3075.
-
(2007). Distinct roles of Wnt/β-catenin and Bmp signaling during early cardiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2007 Nov , 104 (47):18531-18536.
-
(2007). Wnt/β-Catenin Signaling and Cardiogenesis: Timing Does Matter. Developmental Cell. 2007 Jul , 13 (1):10-13.
-
(2006). Mesoderm progenitor cells of common origin contribute to the head musculature and the cardiac outflow tract. Development. 2006 May , 133 (10):1943-1953.
Members
Members

Prof. Eldad Tzahor

Dr. Rachel Sarig














Postdoc
Karen Weisinger - kweisinger2@gmail.com
Gabriele D'uva - duva.gabriele@gmail.com
Miriam Ivenshitz - miriam.ivenshitz@gmail.com
Vardina Bensoussan Trigano - vardina.trigano@gmail.com
PhD
Michal Milgrom-Hoffman - michalmilgrom@yahoo.com
Libbat Tirosh - libbatt@gmail.com
Itamar Harel - itamarharel1980@gmail.com
Inbal Michailovici - inbalit11@gmail.com
Arial Rinon - ariel.rinon@gmail.com
Elisha Natan - elishanatan@gmail.com
Adam Wasserstrom - adam.wasserstrom@gmail.com
MSc
Ayelet Levit - ayeletshamai@gmail.com
Elik Chapnik - elikcha@gmail.com
Hadar Hay - hadar.hay2@gmail.com
Roni Winkler - roni.winkler@weizmann.ac.il
Adi Neufeld - adi.neufeld@yahoo.com
Amir Monovich
Hadas Elhanany-Tamir
Shlomi Lazar
Galleries
Contact

Prof. Eldad Tzahor
Weizmann Institute of Science
Department of Molecular Cell Biology
Wolfson Building
Rehovot, 76100, Israel
Office: +972-8-934-3715
Mobile: +972-50-510-9606
Email: eldad.tzahor@weizmann.ac.il
Find us on campus, Building 33.