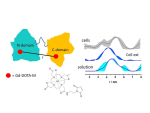
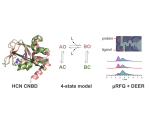
New spin labeling approaches
Electron paramagnetic resonance (EPR) techniques, specifically double electron-electron resonance DEER (or pulsed electron double resonance, PELDOR) are highly effective for determining the distances between two strategic sites in biomolecules such as proteins, nucleic acids, and their assemblies. These are usually carried out in frozen solutions and provide, for example, sparse structural information that can be used for tracking conformation changes upon ligand/substrate binding. At the heart of this methodology lies the controlled labeling of the molecules of interest with paramagnetic probes, between which the distances are measured. These could be either intrinsic paramagnetic centers such as transition metal ions and radicals or artificially introduced spin labels.