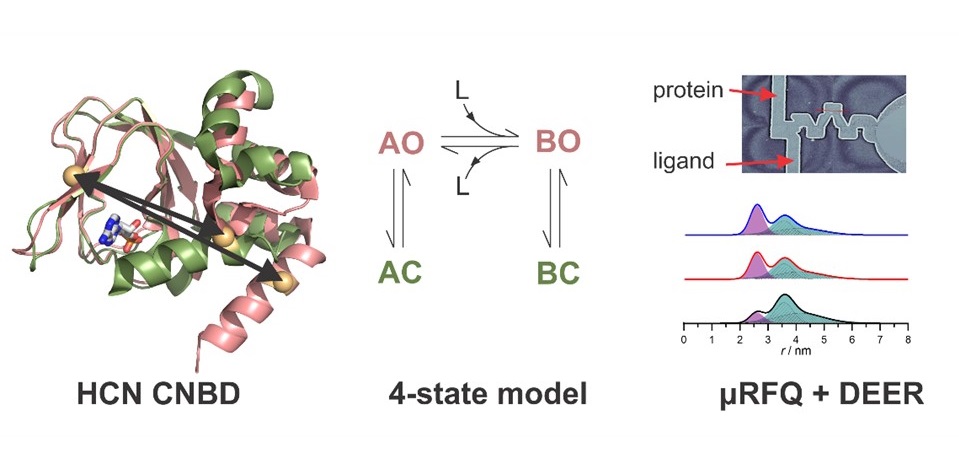
Many proteins undergo large structural changes as they function. X-ray crystallography and cryo- electron microscopy offer detailed snap-shots of single states, but alternative methods are necessary to capture the range of structures occurring in solution. By affording accurate measurements of the distance between two paramagnetic centers, EPR spectroscopy in combination with suitable tags offers unique opportunities to investigate the structural changes that proteins can undergo. In this context, we are interested in trapping intermediate conformations that appear during the protein’s function and are important for elucidating the mechanism of the protein/enzyme. For this purpose, we have developed a microfluidic freeze quench system that is designed for trapping samples for W-band pulse EPR measurements on a millisecond time scale, minimizing the amount of sample needed.
References
-
Collauto A., Deberg H. A., Kaufmann R., Zagotta W. N., Stoll S. & Goldfarb D. (2017) Physical Chemistry Chemical Physics. 19, 23, p. 15324-15334. DOI: 10.1039/c7cp01925d