Developmental transcriptomics of mushroom body neurons and astrocytes
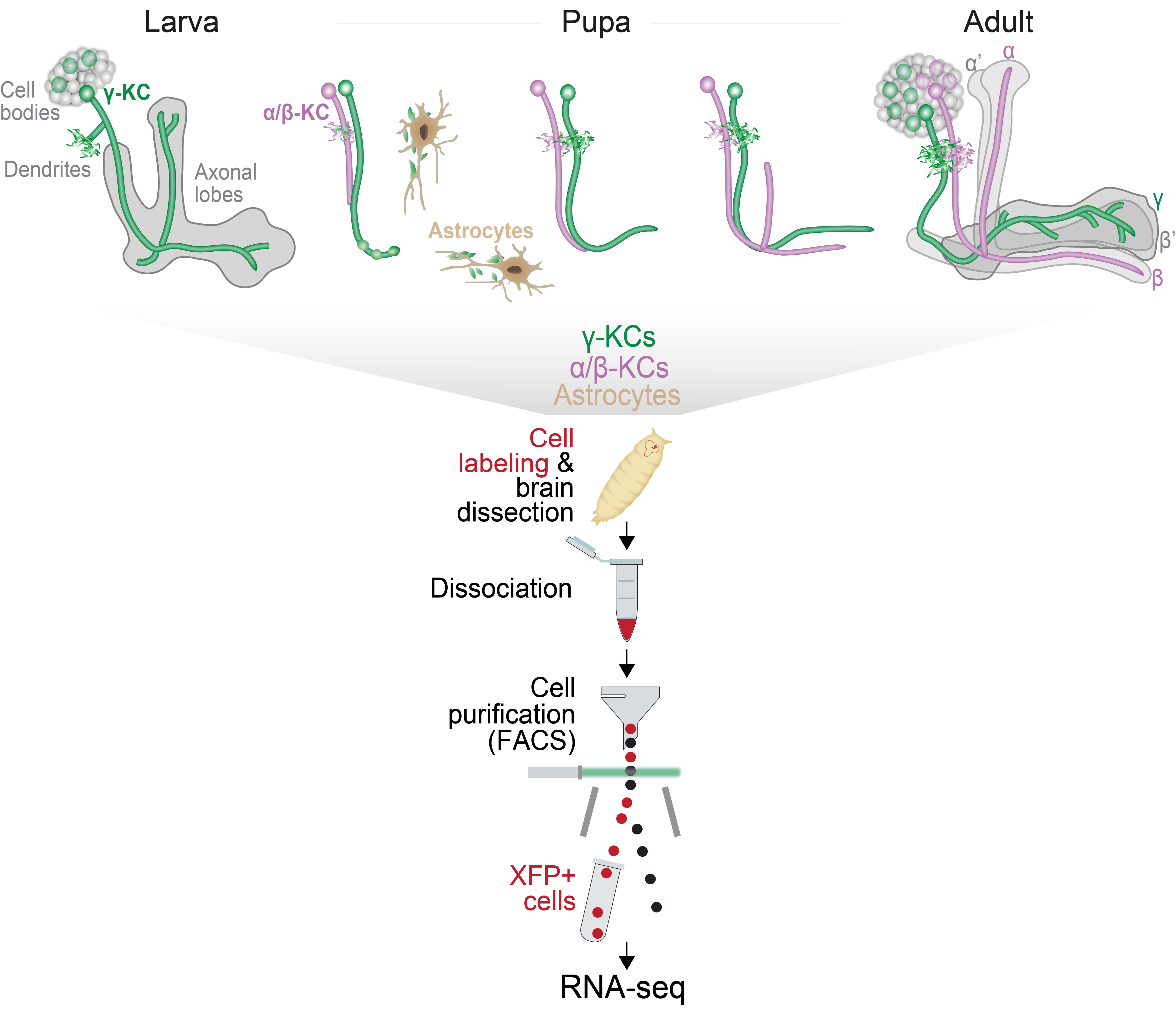
Mushroom body (MB) remodeling is regulated by nuclear receptors, which function as ligand-dependent transcription factors, indicating that these processes are governed by genetic programs. To uncover these programs, we used sorting by FACS followed by RNA-seq to generate expression profiles of MB γ-Kenyon cells (γ-KCs) at 14 developmental stages, using GMR71G10-GAL4 driving the nuclear reporter RedStinger, along with adult MB α/β-KCs (NP3061-Gal4) and adult non-MB neurons (c155-GAL4 additionally expressing MB247-GAL80). In the perturbation-seq experiment, we compared the expression profiles of WT γ-KCs to those perturbed for key regulators of remodeling, including EcR (EcR-DN), Sox14 (Sox14 RNAi), E75 (E75 RNAi) and unf (unf RNAi), at several developmental stages (see Alyagor et al., 2018).
Recently, we expanded our transcriptional profiling of MB α/β-KCs to include 5 developmental stages along their axon outgrowth using the GMR16B01-Gal4 driver (see Fahdan et al., 2026).
Additionally, we profiled the transcriptional landscape of astrocytes in 3rd instar larvae (L3) and in adult brains, using alrm-Gal4 (see Marmor-Kollet et al., 2023).
Our data, as provided below, enables a quick exploration of the expression pattern of any gene in the different developmental datasets. Note that genes which are expressed below a certain threshold are not shown. Please explore our papers for more details.
