The RUNX3 tumor suppressor gene conundrum: An anatomy of a research fraud
(Information supplementing Groner reply to Dr. Ito’s rebuttal-correspondence published in EMBO Molecular Medicine (EMM, June 2012)
Introduction and summary
The rebuttal-correspondence by Dr. Ito (EMM June 2012), addressing the findings described in Levanon et al 2011 EMM article, is nothing but sand thrown in the eyes of readers. As space limitations dictated by Journal policy allowed only brief address of the issues raised in Dr. Ito’s correspondence we herewith offer a more comprehensive account of the major points pertaining to Dr. Ito’s correspondence and to the conundrum regarding Runx3 role in gastric and colorectal carcinogenesis.
The Levanon et al 2011 report challenges data published in Li et al (2002) Cell paper Fig.1E, which showed dark LacZ stained gastrointestinal tract (GIT) of a mouse in which β-gal gene was knocked into the Runx3 locus (Li-Runx3-LacZ mice), in striking contrast to wild-type GIT (Author reply-Fig. 1A). In an attempt to divert the discussion from its main issue, Dr. Ito ignores our comments demonstrating that the data he published in Li et al (2002) Cell paper Fig. 1E, is flawed. Instead he (Dr. Ito) refers the reader to another panel of Fig. 1 (panel I) and to human data not relevant to Li et al 2002 paper. It is important to note that the erroneous data of Fig.1E comprises the foundation of Li-2002 report and its conclusion therein that Runx3 is a tumor suppressor gene (TSG) involved in GIT cancer.
Dr. Ito’s comments in the rebuttal-correspondence also fail to explain why in Rehovot his original Li-Runx3-LacZ mice show no LacZ in GIT while efficient LacZ staining is detected in the known Runx3-expressing sites. Moreover, the LacZ in the other organs is comparable to the originally reported levels both in Li et al and few years later in Yoshida et al (2004). Our failure to detect LacZ in GIT of Li-2002-LacZ mice documented in Levanon et al 2011, and the current inability of Dr. Ito to reproduce his 2002 data, is almost certainly due to the fact that Runx3 was never expressed in GIT epithelium of the Li-Runx3-LacZ mouse described in 2002.
In fact, as documented below, Dr. Ito has previously alluded, more than once, to the potential incorrectness of the data published in Li et al 2002 Cell paper.
1) Dr. Ito alludes to the potential incorrectness of his published 2002 Cell data and admits that he is unable to reproduce the LacZ GIT staining in his Li-Runx3-LacZ mice.
On July 7 2010 Dr. Ito sent the following letter addressed to Yoram Groner (YG) and to the RUNX Forum (17 RUNX leading investigators):
“Dear Yoram:
Thank you for your e-mail, informing me some results of your analysis of our Runx3 KO mouse. I value your rigorous studies on the expression of Runx3 in the GIT epithelium, since your tough comments make us more careful in our analyses.
First, you mentioned that LacZ staining pattern of 14.5 embryo in Fig 1E of our Cell paper published in 2002 is not reproducible. As shown in the Figure, it was Runx3-/- embryo. You analyzed Runx3+/- embryo in which lacZ is a half dose compared with homozygous mouse. We sent you two pairs of heterozygous male and female, so that you can obtain homozygous mice easily. In any case, this point was extensively discussed in Gettysburg. This figure [Li et al Fig. 1E] was prepared when we were still at early days of learning LacZ staining of mouse GIT and, therefore, the figure might not represent the correct Runx3 expression pattern. We will re-examine this point and, if we find it necessary to make a correction, we will do so”.
As documented below Dr. Ito re-examined this issue only a year and a half later and did nothing to correct this fault.
2) More recently it became publically known that Dr. Ito himself is unable to reproduce the fundamental results of his report. This significant development evolved in several steps:
Step-I: The data published in Levanon EMM 2011, including analysis of Li-Runx3-LacZ mice, was presented by our group in July 2010 during the RUNX annual meeting in Hiroshima; first, privately to Dr. Ito and subsequently in the meeting’s formal session.
Step-II: Eight months later, Dr. Ito sent another letter, to YG and to the RUNX Forum reporting that he re-analyzed the Li-Runx3-LacZ mice and, “like Groner”, he is unable to detect LacZ in GIT of these mice [see below clause-1]. To explain these new findings, Dr. Ito first claimed that overall LacZ expression deteriorated, during 10- years of breeding, and the reduced β-gal activity affected LacZ detection in GIT. To support this explanation, Dr. Ito appended a picture showing weaker LacZ staining compared to the original level reported in 2002 (see below Dr. Ito’s letter from March 28 2011 and Fig. 1 B&C herein). However, in striking contrast, analysis of these Li-Runx3-LacZ mice in Rehovot, revealed LacZ levels in the known Runx3 expressing tissues comparable to the original (Cell 2002) levels of Li-Runx3-LacZ mice (Levanon et al, 2011, Fig.6; Fig. 1D herein and Yoshida et al, 2004, Fig.1). Hence, this cannot explain the lack of GIT LacZ in Li-Runx3-LacZ mice.
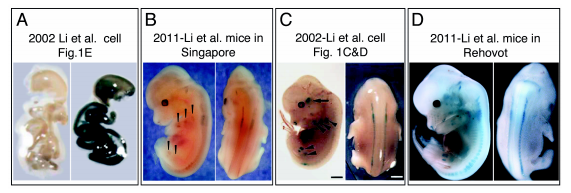
Fig. 1 Analysis of the original Li et al 2002 mice either in Singapore or in Rehovot failed to reproduce the Li et al 2002 data.
A. LacZ staining of E14.5 GIT of Runx3-LacZ mice (right) compared to WT mice (left) (Li et al., 2002, Fig. 1E).
B. E12.5 Li-Runx3-LacZ mice stained in 2011 in Singapore. Lateral and dorsal views show reduced, but clearly visible LacZ staining in DRG compared to the LacZ level published in the Li et al 2002 report shown in C.
C. E12.5 Li-Runx3-LacZ mice stained and published in 2002 (Li et al., 2002, Fig. 1C,D). Shown are lateral and dorsal views. D. E12.5 Li-Runx3-LacZ mice stained in Rehovot in 2011. Shown are lateral and dorsal views, demonstrating comparable LacZ level to the original 2002 level shown in C. Hence, no reduction in LacZ staining of Li-Runx3-LacZ mice is observed.
Step-III: A further, more recent development, Dr. Ito concedes that he is unable to reproduce the 2002 GIT staining because the LacZ expression has completely disappeared: “the marker gene [β-gal] could have been deleted or silenced” (Normile, 2011).
Step-IV: Even more interesting is Dr. Ito’s claim in EMM Correspondence Fig. 2; a breeding schedule-mediated exon-skipping, which “explains why Levanon could not observe LacZ staining in GIT epithelium” [of Rehovot-bred Li-Runx3-LacZ mice]. However, all RT-PCR signals shown in Dr. Ito’s EMM correspondence Fig. 2 (WT and exon-skipping) are most probably contaminants resulting from improper usage of only partially purified GIT epithelium [see below clause-2].
The p33 issue including Dr. Ito’s comments in the correspondence regarding the article by Raveh et al, (2005) were previously addressed by Dr. Gat, the PI and senior author, in response to Dr. Ito’s letter to the RUNX Forum [see clause-3].
In summary, using eight different anti-Runx3 Abs, in situ hybridization, TaqMan qRT-PCR, 3- different reporter-mouse strains (Runx3-GFP-KI, R26-LacZ/Runx3Cre and R26- tdTomato/Runx3Cre) and rigorous re-analysis of the original Li-Runx3-LacZ mice, we are unable to demonstrate Runx3 expression in GIT epithelium although it is readily demonstrable in a series of adjacent tissues. Thus, the inevitable conclusion is that normal GIT epithelium lacks Runx3 expression and the Li-2002 report, which indicated expression of Runx3 in this epithelium, is flawed.
Accordingly, our failure to detect LacZ in GIT of Li-2002-LacZ mice and the current inability of Dr. Ito to reproduce his 2002 data is almost certainly due to the fact that Runx3 was never expressed in GIT epithelium of the Li-Runx3-LacZ mouse described in 2002.
Supplemental information to Groner Response: Clause-1
Data presented in Li et al 2002 Figure 1E could not have been obtained as actual experimental results under any condition of LacZ staining
Depicted below are two panels providing compelling evidence that the data of betagalactosidase (β-gal) staining in GIT of Runx3 KO mice in the original Li et al 2002 Cell paper could not have been produced as claimed and is fraudulent. The enclosed panel A reproduces the images of the Cell paper Figure 1E. Thus, the GIT of a wildtype (WT) mouse is displayed on the left, while the GIT of a Runx3 -/- (homozygous knockout) Li et al mouse is displayed on the right. In the Li et al Cell paper these two samples were said to have been stained for β−gal activity (LacZ).
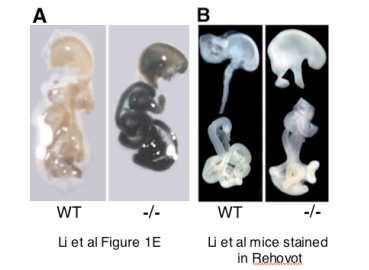
As is apparent in panel A, the WT GIT of the original Li et al 2002 mice, did not stain at all while the homozygous knockout GIT stained dark blue/black. Panel B shows LacZ staining of the GIT of Li et al mice that were provided to us by Dr. Ito and stained in our laboratory with LacZ staining reagents. In panel B, the GIT staining intensity in Li et al Runx3-/- and the WT mice is identical and shows no evidence whatsoever of LacZ staining in Runx3-/- GIT. As the images in panel B provide clearcut evidence for the absence of β-gal activity in GIT of the original Li-mice, the inevitable conclusion is that Li et al 2002 Figure 1E was fabricated. As has been noted above, this irreproducible dark GIT staining shown in Li et al 2002 Figure 1E has more recently disappeared in the original Li-Runx3 mice bred in Dr. Ito’s lab in Singapore. See below the relevant parts from Dr. Ito’s March 28 2011 letter, addressed to YG and “RUNX Forum” and March 29 YG answer.
“28 March 2011
Dear Yoram:
Expression of LacZ in DRG of our current Runx3 KO mice is very weak.
Last year [July 2010] in Hiroshima, you showed us pictures demonstrating the absence of LacZ staining in the GIT epithelium of the Runx3 knockout mouse that we sent you. We were very concerned and, therefore, did the same experiment in my lab. To my surprise, we obtained similar results to yours. Then we examined the expression of Runx3 in DRG in Runx3+/- E12.5 (FIG. A). Earlier, we reported the result shown in FIG. B (Fig 1C,D of Li et al, 2002). We now find very weak LacZ staining of the DRG. The results were reproducible. When we observe the embryo from the back, we can recognize weak expression of LacZ in the DRG. When we take a lateral view, the LacZ staining is rather obscure (FIG. A). As both our groups have previously reported, blue color was also visible in tissues like cartilage in the arms and legs. However, the currently available Runx3 KO mice in my lab show almost no staining. Therefore, I assume those that we sent to you would be the same. If you carefully observe, you may be able to discern barely visible staining of cartilage in arms and legs (indicated by arrowheads in FIG. A). The staining in this embryo is the 5 strongest of the three embryos we examined in this most recent experiment. Under such conditions, it would be quite unfeasible to detect blue color staining in the GIT epithelium. It seems that the expression of the non-functional Runx3-LacZ fusion transcripts has been gradually diminishing in the later generations of our Runx3 knockout mice. I have heard from other investigators who experienced similar kind of loss of expression. I assume you must be obtaining essentially the same results when you analyzed our Runx3 KO mice.
Although I did not expect this to have happened and I expected to see blue color in GIT epithelium, I apologize that I forced you to do the experiments that turned out to be a waste. I am deeply sorry about this”.
“March 29 2011
Dear Yoshi,
Thank you for your kind note of March 28 and the information. I am puzzled by the content of this letter and wonder what motivated you to send it, because I know that you know that what is written in this letter is completely incorrect.
More then eight months ago, at the Hiroshima RUNX meeting, we (Ditsa and I) set with you in private, as an act of courtesy and respect to you, before I made my formal presentation. During this friendly interaction we showed you all the data in great details. You asked questions and we answered.
During these deliberations I have indicated to you that the intensity of DRG and cartilage staining of the Li et al-Runx3 KO mice that you sent us, was as published in Li et al paper and that the only irreproducible data was the dark LacZ staining of the GIT compared to the completely white WT GIT. I have noted that this GIT LacZ staining in Li et al report was ~100 times stronger than the staining of the DRG and cartilage shown it Li et al Figure 1C&D. (See enclosed Fig. 1 and Fig. 2).
I have pointed out to you during this friendly discussion that based on our data it is my conviction that the dark LacZ staining in the GIT could not have been obtained as an actual experimental result. You then responded by saying that Kosei Ito conducted this experiment”.
Conspicuously, the intensity of LacZ in the Li-Runx3-LacZ mice bred in Singapore deteriorated gradually from “strongly expressed” (2002) to “very weak” March 2011 until “completely disappeared” Oct 2011. In parallel, the “exon-skipping” explanation described in Dr. Ito’s EMM correspondence emerged. For example, this “exon-skipping” notion was not mentioned in “the long rebuttal [document] provided to Science” on Oct 2011 (Normile 2011).
Supplemental information to Groner Response: Clause-2
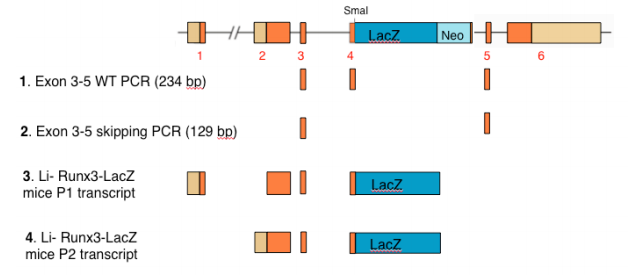
In an attempt to find a pretext to his current failure to reproduce the data published in Li et al 2002 Cell report, which show high expression of Runx3-LacZ in mice GIT, Dr. Ito provides in his EMM correspondence a new explanation; the continuous breeding of the Li-Runx3-LacZ mice evoked “exon skipping” which eliminates Runx3 exon 4 including the LacZ-neo cassette (see scheme below). To support the exon-skipping claim, Dr. Ito included in the correspondence an RT-PCR analysis (correspondence Fig. 2) presumed to demonstrate that “exon 4 and LacZ are absent from a large fraction of Runx3 RNA in GIT epithelium” [of Li-Runx3-LacZ mice]. The experiment and its conclusions are scientifically unsound for the following reasons:
1) In clear contradiction to the data published in Li et al 2002 Cell paper that: “Strong β-gal activity was found in gastrointestinal organs, including stomach and small and large intestines from 14.5dpc through to adulthood”, Dr. Ito’s correspondence claims that now they find that “the level of cellular Runx3 RNA in mouse GIT is low (Fig. 2)”. This new finding is based on RT-PCR, which was conducted using RNA isolated from partially purified GIT epithelium (correspondence Fig. 2 legend). Given that the GIT is heavily populated by tissue embedded leukocytes expressing high levels of Runx3, the validity of the data presented in Fig. 2 is doubtful. In fact, by a more rigorous approach entailing usage of FACS sorted GIT epithelial cells and TaqMan primed qRT-PCR we have demonstrated the complete absence of Runx3 RNA in WT GIT epithelium (Levanon 2011 EMM Fig. 5L). Thus, the Runx3 and Runx3-LacZ RT-PCR signals in Dr. Ito's correspondence Fig. 2, are most probably due to contaminating GIT leukocytes.
2) The use of partially purified GIT epithelium in Fig. 2 experiment of Ito’s correspondence invalidates the presented data for yet another reason. The Li-Runx3- LacZ mice contain a large 5.5 kb LacZ-neo cassette inserted into the small (0.1 kb) exon 4 (see scheme below). Based on the conventional wisdom under such circumstances, the majority of transcripts will terminate following the LacZ poly-A signal and only a minor fraction will extend into the neo cassette-exon 5. Hence, in tissues of Li-Runx3-LacZ mice that genuinely express Runx3, including GIT leukocytes, the expression of LacZ is mediated by the P1 and/or P2 transcripts shown below (i.e. numbers 3 and 4 in the scheme). As evidenced by the comprehensive analysis described in Levanon 2011 EMM report, Runx3 is not detected in GIT epithelium. Thus, the RT-PCR products shown in Dr. Ito’s correspondence Fig. 2, must have resulted from the minute amount of WT leukocyte Runx3 RNA (Fig. 2 three left samples) or exon skipped Runx3-LacZ RNA (three middle samples) of GIT leukocytes contaminating their partially purified samples (correspondence Fig. 2 legend). As no attempt was made by Dr. Ito et al to quantitate the relative abundance of the RTPCR products, the data in Fig. 2 merely provides inconclusive information about GIT leukocyte Runx3 RNA of WT and Li-Runx3-LacZ mice.
3) In a recent analysis performed in Dr. Ito’s lab in Singapore (see above Dr. Ito’s letter from March 28 2011), the Li-Runx3-LacZ mice displayed reduced, but clearly visible LacZ staining in dorsal root ganglia (DRG). Thus, one would expect that correspondence Fig. 2 would include a positive control of DRG RT-PCR analysis demonstrating a Runx3-LacZ transcript in which exon 4 was not skipped. In fact, a more recent analysis of Li-Runx3-LacZ mice conducted in Rehovot (Figure 1 panel D), clearly demonstrated that the current LacZ level in DRG and cartilage of Li- mice is comparable to that originally shown in Li et al 2002 Cell report. This finding refutes the claim that a breeding-schedule mediated exon-skipping phenomenon affecting a large fraction of Runx3-LacZ RNA has occurred in Li-Runx3-LacZ mice.
Taken together the impurity of the GIT epithelium and improperly conducted RT-PCR analysis render the data presented in EMM correspondence Fig. 2 non-informative and misleading.
Scheme showing the Runx3 gene (top), the predicted size of RT-PCR products 1 and 2 between exons 3-5 and the predicted Runx3-LacZ RNAs 3 and 4.
Supplemental information to Groner Response: Clause-3
In the EMM correspondence Dr. Ito raises the question as to why they [Levanon et al] analyze mainly mouse tissues and thereafter discusses the non-relevant issue of Runx3 p33 isoform. As was previously noted, Levanon et al 2011-EMM report specifically challenges data published in Li et al (2002) Cell paper Fig.1E, which showed dark LacZ stained GIT of the Li-Runx3-LacZ mouse, in striking contrast to wild-type GIT (see Li et al 2002 Cell 109, 113-124 or Groner Response-Fig. 1A). The data reported in Levanon et al EMM 2011 refute the results published in Li 2002 Fig. 1E as well as the paper’s take-home message and demonstrate that the published Li2002 data could not conceivably have been produced as claimed in the Li-2002 manuscript. Accordingly, the current inability of Dr. Ito to reproduce his 2002 data is almost certainly due to the fact that Runx3 was never expressed in GIT epithelium of the Li-Runx3-LacZ mouse described in 2002. Bluntly, Li et al Cell Fig. 1E was fabricated.
It should also be noted that the anti-Runx3 antibodies (Ab) R3-1E10, which were raised and extensively used by Dr. Ito and other investigators to demonstrate Runx3 expression in GIT epithelium has aberrant characteristics (Levanon et al 2011). These deviant features, which were also noted by Dr. Ito, indicate that R3-1E10 cannot be considered specific and thus is not a valid anti-Runx3 Ab. As such, we believe, it is scientifically unsound to use R3-1E10 without proper disclosure of information regarding its unique properties, as was done in the Ito et al, 2008 report (Ito K. et al Cancer Cell 14, 226-237).
Concerning p33 Runx3 isoform. As already noted, the p33 bears no relevance to the problematic data in Li et al Cell paper and to the challenge of this Li et al 2002 data by Levanon et al EMM 2011 report. The in-length discussion of the p33 issue in Dr. Ito correspondence is merely a “sand thrown in the eyes” tactic aimed at diverting the discussion from the crux of the problem.
The potential existence of human and mouse Runx3 p33 isoforms was first noted by us during cloning and subsequent sequencing of the human and mouse RUNX3/Runx3 genes. In mouse tissues p33 is hardly detected. It is readily detected in long-term cultured bone marrow derived dendritic cells and in the cultured dendritic cell line D1. However, it is not detected in GIT leukocytes. The p33 protein lacks a major part of the DNA binding domain characterizing the RUNX family (the so-called runt domain) and its biological significance and in vivo function are not known. For example, we have previously generated mice lacking the ability to produce p33 by crossing floxed Runx3 exon 3 and transgenic-Pgk-Cre mice. Analysis demonstrated that homozygous Runx3exon3-/- mice display all the known features of Runx3-null mice and thus are phenotypically indistinguishable from the other Runx3 KO mouse strains.
Attempting to justify the discussion over p33, Dr. Ito refers in his correspondence to the paper by Raveh et al (Dev Dyn. 2005, 233 1478-87) in stating: “Indeed, the lack of LacZ expression in the Runx3-positive epidermal cells in Rehovot-Runx3 KO was reported (3rd paragraph in p1481, Raveh et al, 2005)”. As Dr. Ito has already raised and discussed this issue in a letter (dated March 31 2011) to the RUNX Forum and while Levanon and Groner are co-authors of Raveh et al 2005, the senior author of this report is Dr. Uri Gat whose laboratory at the Hebrew University, conducted all the experiments mentioned in Dr. Ito correspondence. Thus, Dr. Gat is the most qualified person to respond to comments regarding the Raveh et al 2005 report.
Enclosed below is his original answer to Dr. Ito and the RUNX Forum, which speaks for itself.
On 4/7/11 11:21 PM, "Dr. Uri Gat" wrote:
Dear Yoshiaki,
Yoram and members of the Runx research community, I would like to share with you the status of Runx3 expression in skin, following Yoshiaki's letter of March 31. You are all welcome of course to re-read our Dev Dyn 2005 paper, in which we have first explored Runx3 expression in mouse skin and the Runx3 KO hair phenotype. The May 31 letter described an excerpt from the paper about the epidermal expression, which may be misleading. Most (>95%)of the staining in skin is in the dermal papillae of the hair follicle (based on several antibodies, β-galactosidase and in situ hybridization) that is mesodermal in origin. A minority of the staining in the hair follicle is of melanocytes, which are neural crest in origin. In the epidermal compartment there is a small percentage of isolated (nonepithelial) Runx3 positive cells (β-gal negative), which are most likely dendritic cells but may be other immune cells. These cells however are NOT Keratinocytes and are NOT epithelial. Thus these isolated cells that just reside in the epidermis cannot be likened in any way to the epithelial cells of the GIT. They are different in their embryonic origin and in their tissue type, which is single migratory cells and not epithelial cells. The mere finding that some cells in a mouse model express β-gal 9 while others (totally different) do not, is of no real contribution to an argument where the same tissue (even in the same mouse model) is positive in one lab and negative in another using several different methods of detection. These are the facts regarding the expression we have found in the haired skin - below please find the relevant excerpt from our paper.
I would like to use this opportunity to address a related important issue about expression of Runx3 in skin that has bothered me for a long time but I did not have the opportunity to deal with. This has to do with human skin and I'd like to state that we have stained normal human skin with the same polyclonal Runx3 antibodies and have found the expression pattern quite similar to that of mouse. This is in contrast to the results published by Salto-Tellez et al. (Oncogene (2006) 25, 7646–7649), in which Ito is the corresponding author. In this paper Runx3 is shown to be highly expressed in most cells of the normal human epidermis in cells that are evidently epithelial keratinocytes (Salto-Tellez et al. Fig. 2 b and d). The dermal cells are also highly stained - something which we have never seen as well. Most strangely, in a western blot of normal skin cells, which appears in the next figure, this high level staining is not reflected, since there are no bands! (please check!). Thus, I conclude that the results shown in this epithelial tissue (human epidermis) are an artifact. At least in skin whether murine or human there is no epithelial Runx3 expression. I hope this account can clear the skin expression issue and perhaps also contribute to this ongoing debate. All the best, Uri .
----------------------------------------------------
Uri Gat, Department of Cell and Developmental Biology
The Silberman Life Sciences Institute
Room 3-575 Edmond Safra
Campus in Givat-Ram
Hebrew University in Jerusalem Israel, 91904
Tel/Fax: (972)-2-6585920
E-mail: gatu@vms.huji.ac.il
References
Ito K, Lim AC, Salto-Tellez M, Motoda L, Osato M, Chuang LS, Lee CW, Voon DC, Koo JK, Wang H, Fukamachi H, Ito Y (2008) RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell 14(3): 226-237
Levanon D, Bernstein Y, Negreanu V, Bone KR, Pozner A, Eilam R, Lotem J, Brenner O, Groner Y (2011). Absence of Runx3 expression in normal gastrointestinal epithelium calls into question its tumour suppressor function. EMBO Mol Med. 3(10): 593-604
Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, Yamashita N, Yano T, Ikeda T, Itohara S, Inazawa J, Abe T, Hagiwara A, Yamagishi H, Ooe A, Kaneda A, Sugimura T, Ushijima T, Bae SC, Ito Y (2002) Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109(1): 113-124
Normile D (2011) Cancer research. Dispute over tumor suppressor gene Runx3 boils over. Science 334(6055): 442-443
Raveh E, Cohen S, Levanon D, Groner Y, Gat U (2005) Runx3 is involved in hair shape determination. Dev Dyn 233(4): 1478-1487
Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, Komori T (2004) Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev 18(8): 952-963