Visualizing microbial societies
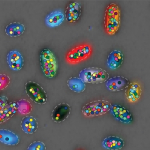
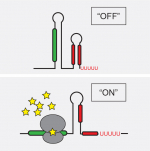
Phenotypic heterogeneity, a form of microbial “individuality” that underlies clonal population survival and sociality, has so far been reported primarily through serendipity. Our lab now seeks to make the study of phenotypic variation systematic and to use this new perspective to gain fundamental insights into its core principles and functional roles. We are employing state-of-the-art single-cell and spatial transcriptomics approaches to realize this goal and shed light on how phenotypic heterogeneity manifests in environmental and host contexts.