Dynamic compartmentalization is a central part of all living systems. In eukaryotes, subcellular mRNA targeting regulates expression levels and enables localized protein synthesis. By contrast, it was long assumed that bacteria lack such RNA-based spatial regulation due to their “minimalistic” cellular architectures. However, strong evidence for RNA localization has accumulated over the last several years in various bacterial species suggesting spatial regulation may be a widespread feature in prokaryotes. However, due to the technical challenges of studying transcriptome organization with subcellular resolution, RNA localization remains largely unexplored.
We recently developed a multiplexed imaging platform based on single-molecule Fluorescence In Situ Hybridization (smFISH) that can now bridge these technological gaps (Dar et al., Science 2021). Our approach, called parallel and sequential smFISH (par-seqFISH), allows us to map the subcellular localization of hundreds of different mRNAs in bacteria grown under diverse conditions. Our lab leverages this technology to acquire a systems biology view of bacterial RNA localization, its targeting mechanisms, functional implications, and evolutionary dynamics.
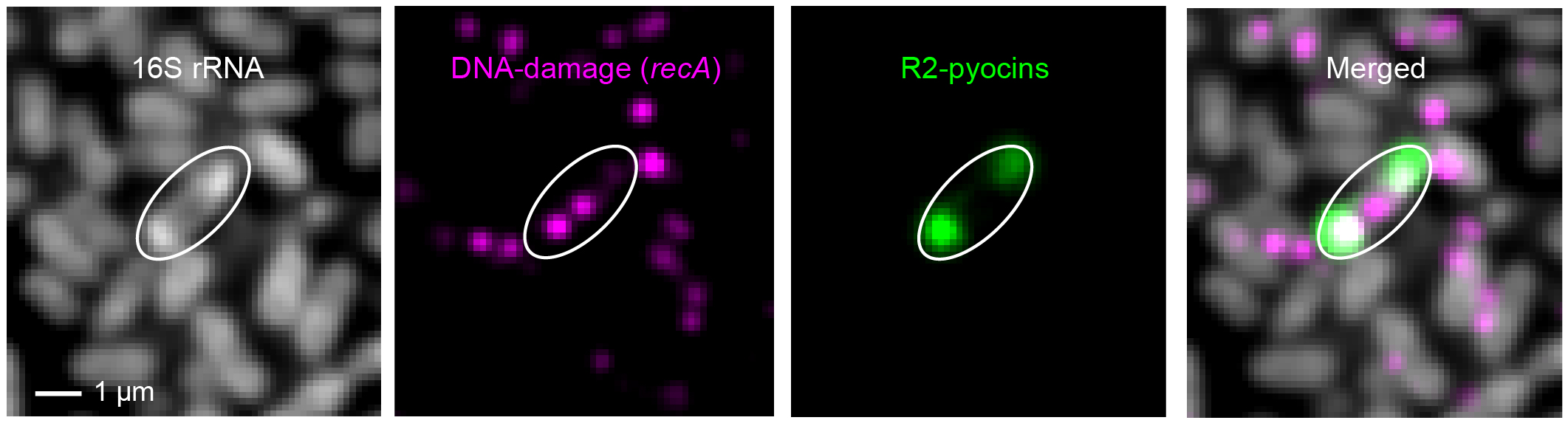
Pyocins are phage tail-like elements that adapted to the host and are now used as secreted antimicrobials. We found that pyocin mRNAs and the cellular ribosomes (16S rRNA) co-localize to the cell poles in Pseudomonas aeruginosa. By contrast, other expressed genes (e.g., recA) are randomly distributed, suggesting a pyocin-specific targeting mechanism. We hypothesize that this compartmentalization increases local translation efficiency and pyocin particle assembly.