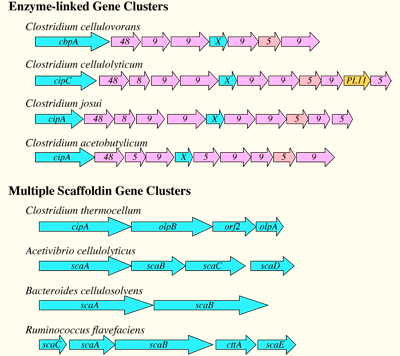
Bacterial cellulosomal systems can be categorized into two major types: simple cellulosome systems contain a single scaffoldin and complex cellulosome systems exhibit multiple types of interacting scaffoldins. The genes encoding for many important cellulosome subunits are organized in “enzyme-linked gene clusters” on the chromosome. In the simple cellulosome systems, the scaffoldin gene is followed downstream by a series of genes that code for dockerin-bearing enzymes. In the complex cellulosome systems, the scaffoldin genes are organized into “multiple scaffoldin gene clusters” on the chromosome as shown in the following scheme:
The Scaffoldin Subunits
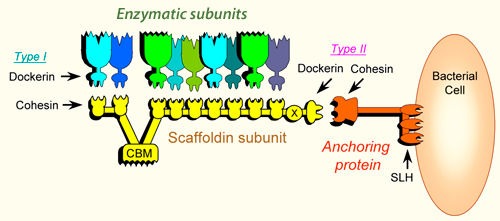
The scaffoldin subunit contains one or more cohesin modules connected to other types of functional modules. In a given scaffoldin, the latter types of modules may include a cellulose-specific carbohydrate-binding module (CBM), a dockerin, X modules of unknown function, an S-layer homology (SLH) module or a sortase anchoring motif. The arrangement of the modules on the scaffoldin subunit and the specificity of the cohesin(s) and/or dockerin for their modular counterpart dictate the overall architecture of the cellulosome. Several different types of scaffoldins have been described: the primary scaffoldins incorporate the various dockerin-bearing subunits directly into the cellulosome complex, adaptor scaffoldins increase the repertoire or number of components into the complex, and the anchoring scaffoldins attach the complex to the bacterial cell surface.
Simple Cellulosome Systems
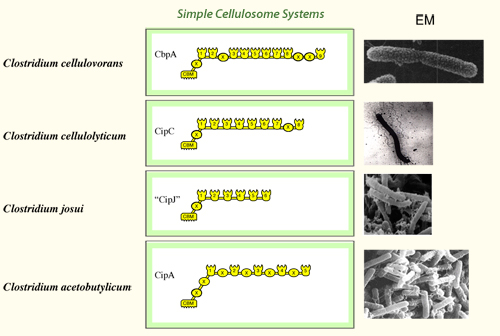
In the simple cellulosome systems, the scaffoldins contain a single CBM, one or more X2 modules and numerous (5 to 9) cohesins. These scaffoldins are primary scaffoldins, which incorporate the dockerin-bearing enzymes into the complex. In several cases, the simple cellulosomes have been shown to be associated with the cell surface, but the molecular mechanism responsible for this is still unclear. The X2 module may play a role in attachment to the cell wall. The scaffoldins of simple cellulosome systems are given in the following scheme:
Complex Cellulosome Systems
In these systems, more than one scaffoldin interlocks with another in various ways to produce complex cellulosome architectures. At least one type of scaffoldin serves as a primary scaffoldin that incorporates the enzymes directly into the cellulosome complex. In each species, another type of scaffoldin attaches the cellulosome complex to the cell surface via a specialized module or sequence, designed for this purpose. Schematic representations of complex cellulosome systems of the following bacteria are shown:
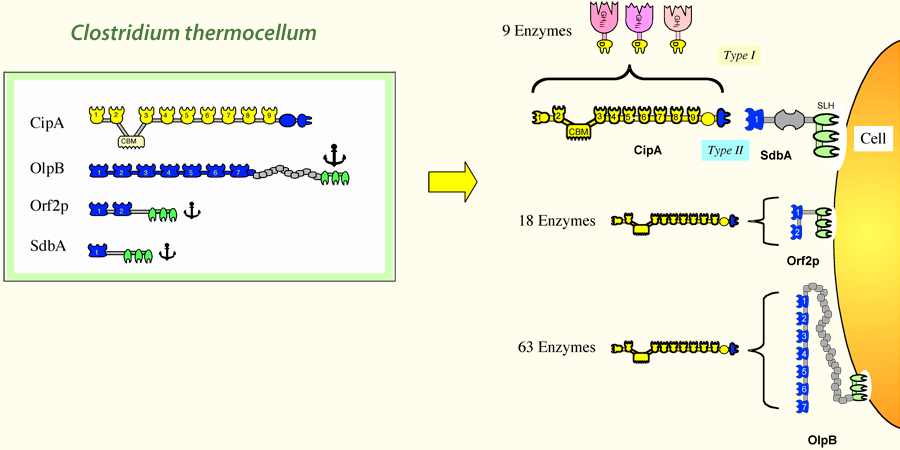
The Clostridium thermocellum cellulosome paradigm consists of individual dockerin-containing enzymes integrated into the 9 cohesins of the primary scaffoldin. A set of anchoring scaffoldins possess 1, 2 or 7 cohesins for attachment of the corresponding numbers of primary CipA scaffoldins and their enzyme complement to the cell surface.
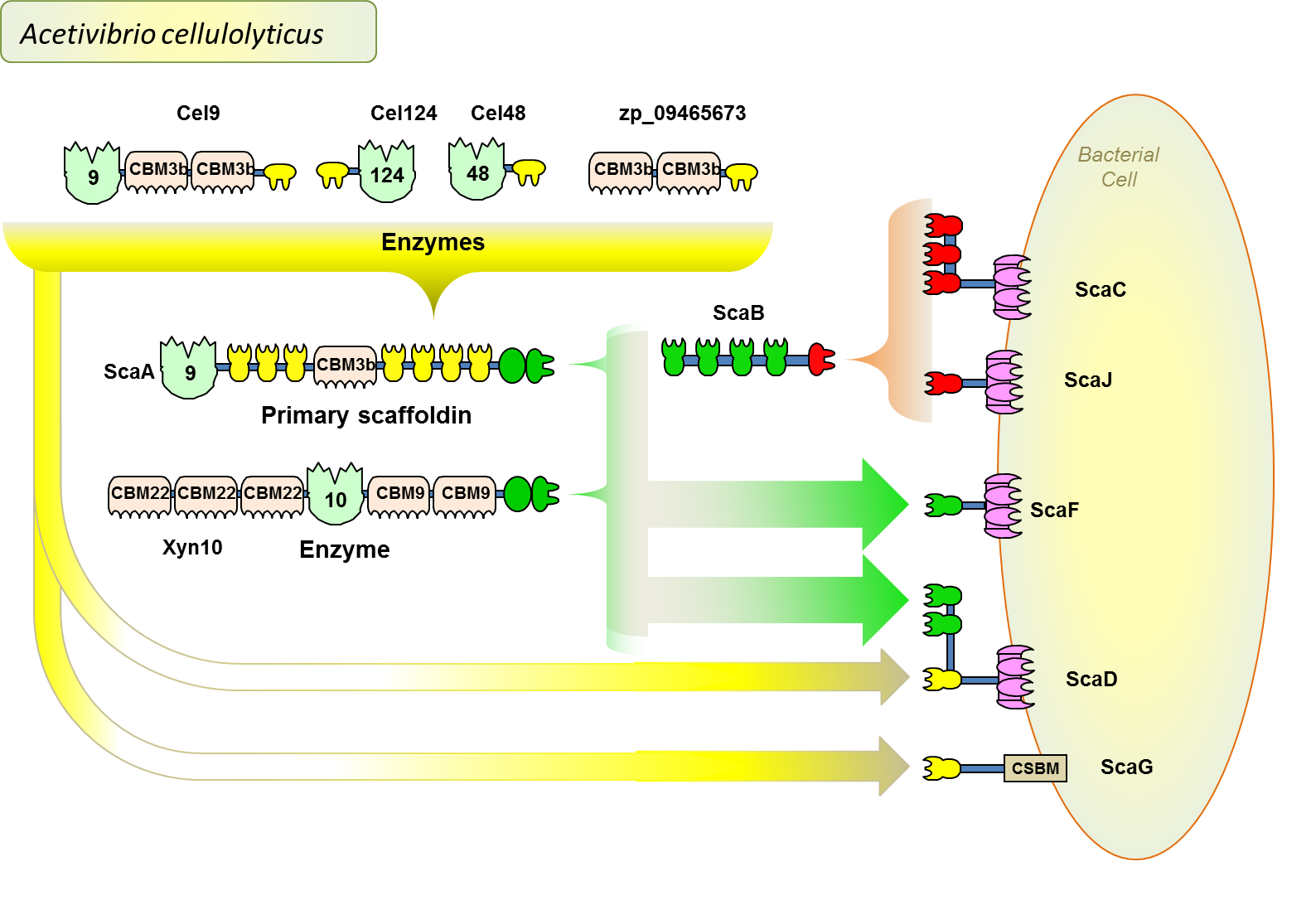
The intricacy of the Acetivibrio cellulolyticus cellulosome system reveals a very complicated cellulosome system — unique from that of C. thermocellum. Here are presented the cellulosomes which are attached to the cell surface by anchoring scaffoldins. One of the cellulosome complexes contains a unique adaptor scaffoldin (ScaB), and another contains a unique bifunctional scaffoldin (ScaD) that plays a dual primary/anchoring role.
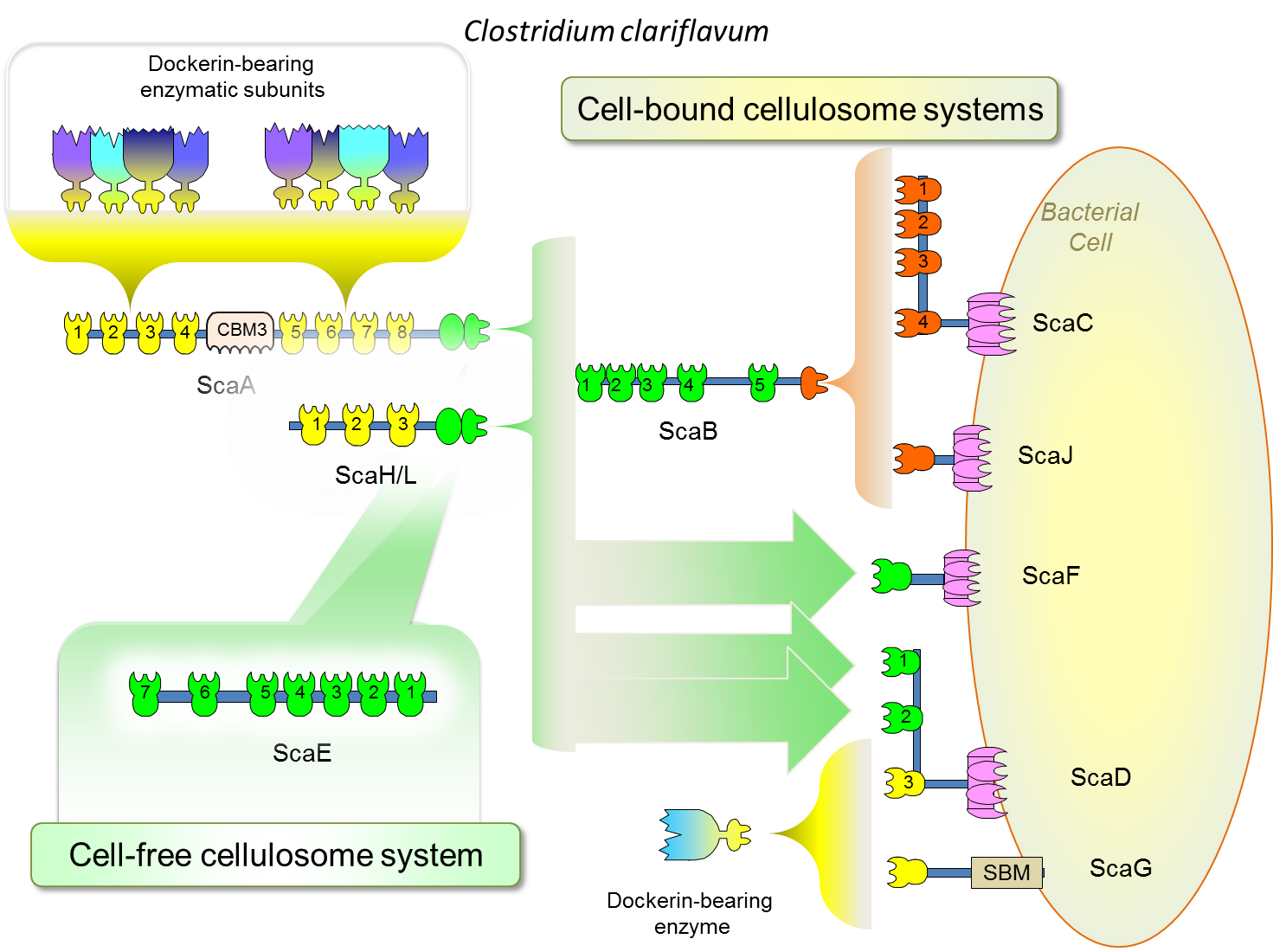
The cellulosome system of C. clariflavum closely resembles the cellulosome system of A. cellulolyticus. C. clariflavum is capable of producing the largest cellulosome complex known in nature, which contains up to 160 enzymes. This complex is composed of the primary scaffoldin ScaA, the adaptor scaffoldin ScaB and the anchoring scaffoldin ScaC. In addition, there are cell-free complexes, assembled onto the ScaE scaffoldin, that can work independently without the presence of the bacterial cells. A similar type of ScaE scaffoldin is characteristic of cellulosome systems from other related bacteria, e.g., C. thermocellum, A. cellulolyticus and B. cellulosolvens.



