The Cohesin module
Cohesin modules are the major building blocks of scaffoldins which are responsible for organizing the cellulolytic subunits into the multi-enzyme complex.
The Dockerin module
The dockerin module anchors the catalytic subunits to the scaffoldin. It displays internal two-fold symmetry, consisting of a duplicated F-hand motif (a calcium-binding loop preceding an helix).
Cohesin-dockerin interaction
The cohesin-dockerin pair represents a high-affinity (Kd 10-9-10-12 M) protein-protein interaction. This interaction tends to be nonspecific within a given species but specific between species. Thus, a scaffoldin-borne cohesin of one species would be expected to recognize and bind to most or all of the enzyme-borne dockerins from the same species, but not with the dockerins from a different species.
Calcium ions are critical to the interaction due to the calcium-binding motif of the dockerin domain whose sequence resembles the EF-hand motif of eukaryotic calcium-binding proteins.
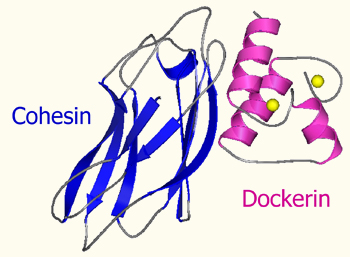
The type-I cohesin-dockerin interaction
Our work focuses on the molecular basis of the interaction between the binding domains of the cellulosome using biochemical and molecular biological techniques. We employed progressive deletion and alanine-scanning techniques on a representative type-I dockerin module from Clostridium thermocellum, which demonstrated that only one of the repeated motifs is critical for high-affinity cohesin binding. The results suggested that the near-perfect symmetry in sequence and structure of the repeated elements of the dockerin is not essential to cohesin binding. Essentially the same conclusion was verified by systematically mutating the highly conserved residues in the calcium-binding loop. Mutations in one of the calcium-binding loops failed to disrupt cohesin recognition and binding, whereas a single mutation in both loops served to reduce the affinity significantly. The results are mutually compatible with recent crystal structures of the type-I cohesin-dockerin heterodimer, which demonstrated that the dockerin can bind in an equivalent manner to its cohesin counterpart through either its first or second repeated motif.
The type-III cohesin-dockerin interaction
In this study, we characterized functionally and structurally a unique, atypical type-III dockerin from the Ruminococcus flavefaciens ScaA scaffoldin (ScaADoc). By using different spectroscopic measurements we discovered that calcium binding to the dockerin induces folding into its secondary structure, by which hydrophobic patches are exposed on the surface of the dockerin. Sequence alignment of the type-III dockerin with a representative type-I dockerin suggested that its putative species-specificity residues are small and hydrophobic. By replacing them (using site-directed mutagenesis) with nucleophilic residues, derived from the type-I dockerin of Clostridium thermocellum, we succeeded in switching the ScaADoc specificity to emulate a type-I cohesin. While the type-I dockerin module exhibits a symmetrical, dual mode of binding to cohesin, the type-III dockerin may bind to cohesin in a different mode, possibly in a non-symmetrical manner and with both segments contributing to complex formation on an equal basis.
Since the mode of interaction of the type-III complex is still unknown, future research will also focus on the structure of the cohesin-dockerin complex by using X-ray crystallography and NMR spectroscopy, in order to further understand the forces that drive this unique tenacious interaction.
Specificity and affinity interplay in the cohesin-dockerin interaction
The very tight interaction between cohesins and dockerins is also known to be species specific, although there is considerable similarity in sequence and structure among cohesins and dockerins of different species. Previous studies in our lab showed that the specificities can be diversified by mutation and/or gene swapping both for the cohesins (Nakar et. al., 2004) and the dockerins (Mechaly et.al., 2001).
In order to broaden our knowledge on cohesin recognition “hotspots” and the relevant residues, we performed a series of single and double point mutations on the cohesin-dockerin interface from the C. thermocellum cellulosome, based on computational analysis of the solved type-I interaction structure. Surprisingly, a single mutation resulted in cross-species binding to a C. cellulolyticum dockerin, however the wild-type interaction was unaffected. Negative design will also be implemented in order to abolish the original affinity.
In the future, we are planning to perform additional manipulations on the interface of various cohesin-dockerin complexes, in order to understand the mechanism of this interaction and its potential for further protein engineering of the cohesin-dockerin interaction. In particular, the work will be done on the less-studied cohesin-dockerin interaction from R. flavefaciens to determine the critical recognition residues, to model the complex and finally to manipulate them by alanine scanning and high-throughput mutagenesis.
Assay systems
Most recently, increasing numbers of cellulosomal and non-cellulosomal cohesins and dockerins are in the process of being discovered, owing to genome sequencing projects. In order to accommodate this tremendous wealth of new cohesin-dockerin interactions, we have developed several complementary systems for broad examination of cohesin-dockerin specificity. These include:
ELISA analysis of the cohesin-dockerin interaction
We have developed expression cassettes that encode two different carrier proteins: (i) a thermostable xylanase with an appended His-tag for expression and purification of the dockerin modules and (ii) a highly stable cellulose-binding module (CBM) for expression and purification of the cohesin modules. Both of these complementary fusion proteins showed very high expression levels of soluble protein in E. coli, thus providing a standardized procedure for there rapid and reliable production. The expressed proteins were extremely stable, and the functionality of the cohesin or dockerin component was retained. The matching fusion-protein systems were used to develop a semiquantitative ELISA system for screening large numbers of cohesin- and dockerin-containing samples.
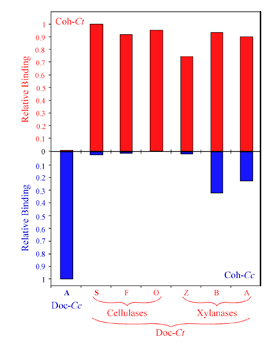
Specificity of C. thermocellum dockerins. Affinity-based ELISA standards were carried out using immobilized CBM-Cohesin constructs from C. thermocellum (red bars) or C. cellulolyticum (blue bars). The C. thermocellum and C. cellulolyticum dockerin fusion proteins (Doc-Ct and Doc-Cc) were used as standards. Additional Xylanase-Dockerin fusion proteins were also produced, comprising the dockerins from C. thermocellum Cel5O (O), Cel9F (F), Xyn10Z (Z), XynB (B) and XynA (A).
Barak et. al., 2005

Example of pcELISA for cohesin mutants.
The xylanase-dockerin fusion protein (Xyn-Doc) was interacted with a concentration gradient of mutant cohesins, prior to interaction with wild-type cohesin-coated wells of microtiter plates (a).
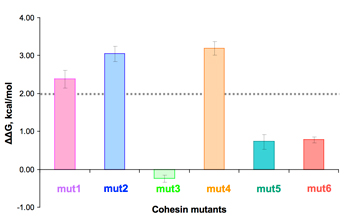
From IC50 values obtained from the plots, the free energy change can be calculated (b). ΔΔG values larger than 2 kcal/mol (dashed line) suggest a significant binding role for the respective mutated residues.
High-throughput microarray system
For these purposes, a matching recombinant fusion protein system was designed, which enables high expression levels of soluble protein and standardization of assay procedures. The cohesins were thus fused to a CBM, and dockerins were fused to a xylanase carrier. These systems allow global comparison among the interactions between various members of these two complementary families of interacting protein modules. Knowledge of the specificity characteristics of native and mutated members of the cohesin and dockerin families provides insight into cellulosome architecture and allows selection of suitable cohesin-dockein pairs for future biotechnological and nanotechnological application.
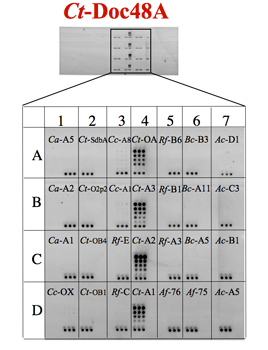
CBM-Coh fusion proteins were printed in triplicates in successive two-fold dilution, and the XynDoc fusion protein was then applied. The amount of dockerin bound to the cohesin samples was visualized by immunofluorscence.
Haimovitz et. al., 2008
Protein affinity purification based on the cohesin-dockerin interaction
A large repertoire of dockerin modules has been assembled in our lab, compiled from dozens of dockerins with varying affinities to cohesin. The collection of dockerins was used as a platform in order to develop a new, affinity-based protein purification system. Several mutated dockerins were chosen in order to evaluate their potential to serve as an affinity tag. Using this approach, we found that a truncated dockerin, DocS (D25), composed of only 47 amino acids, represented the shortest functional dockerin known and could serve as an effective affinity tag. The combination of the relatively small affinity tag, high specificity, and mild elution conditions, provided a “proof of concept” for a new, high-yield, protein purification system.
Single-molecule force spectroscopy (SMFS) studies of the cohesin-dockerin interaction
Mechanical forces play a fundamental role in biological systems. Experimental methods including SMFS allow direct measurement of molecular mechanical properties. As many cellulosomal habitats (for example, cow rumen) exhibit strong flow gradients, and shear forces will accordingly apply mechanical stress on bridging scaffold components in vivo. Protein modules located at stressed positions within these networks should therefore be preselected for high mechanostability. Our group has instituted a collaboration with the group of Herman Gaub and Michael Nash (LMU, Munich, Germany), who are experts in SMFS technology. SMFS experiments were performed to probe the mechanical dissociation of a cohesin-dockerin complex that links the R. flavefaciens cell wall to the cellulose substrate (the cohesin module of the anchoring scaffoldin ScaE (CohE) and its cognate X-dockerin (XDoc) modular dyad from CttA).
The complex was found to withstand forces of 600–750 pN, making it one of the strongest bimolecular interactions reported, equivalent to half the mechanical strength of a covalent bond. Our findings demonstrate force activation and inter-modular stabilization of the complex, and suggest that certain network components serve as mechanical effectors for maintaining network integrity.
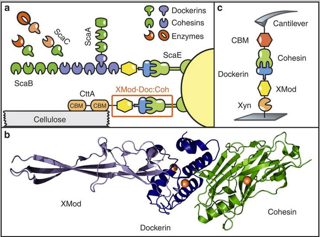
Single-molecule force spectroscopy (SMFS) of an exceptionally strong cohesin-dockerin complex. a) Schematic of selected components of the R. flavefaciens cellulosome. The investigated XMod–Doc:Coh complex responsible for maintaining bacterial adhesion to cellulose is highlighted in orange. b) Crystal structure of the XMod-Doc:Coh complex. Ca2+ ions are shown as orange spheres. c) Depiction of experimental pulling configuration I, with Coh-CBM attached to the cantilever tip and Xyn–XMod–Doc attached to the glass surface.
Steered molecular dynamics simulations were carried out by parallel collaborative studies with Klaus Schulten and Rafael Bernardi (Univ. Illinois, USA). The results revealed that dockerin residues in the binding interface “clamped down” on complementary cohesin residues upon mechanical loading, resulting in increased stability and decreased accessibility of water into the hydrophobic core of the bound complex. These results suggested that a “catch bond” mechanism is responsible for the remarkable stability under force and provided a molecular mechanism which the XMod–Doc:Coh complex employs to summon mechanical strength when needed, while still allowing relatively fast assembly and disassembly of the complex at equilibrium.



