Research
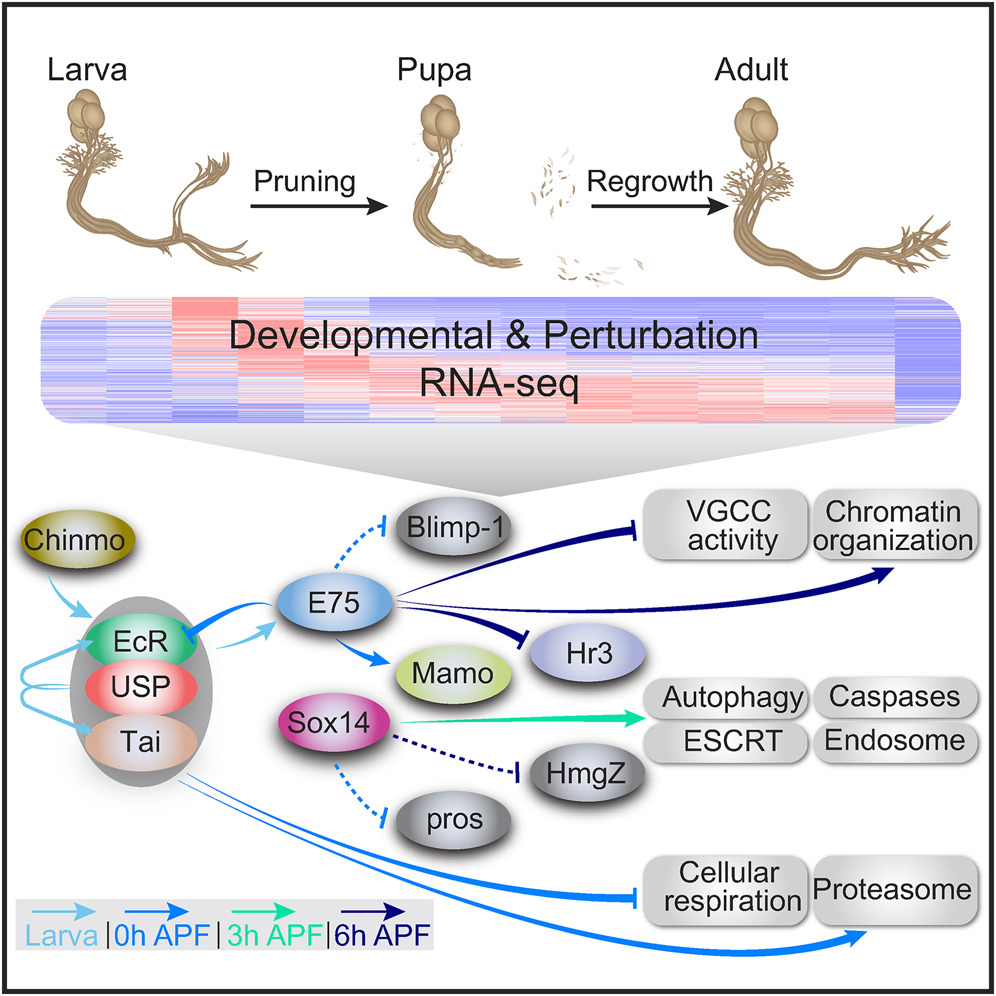
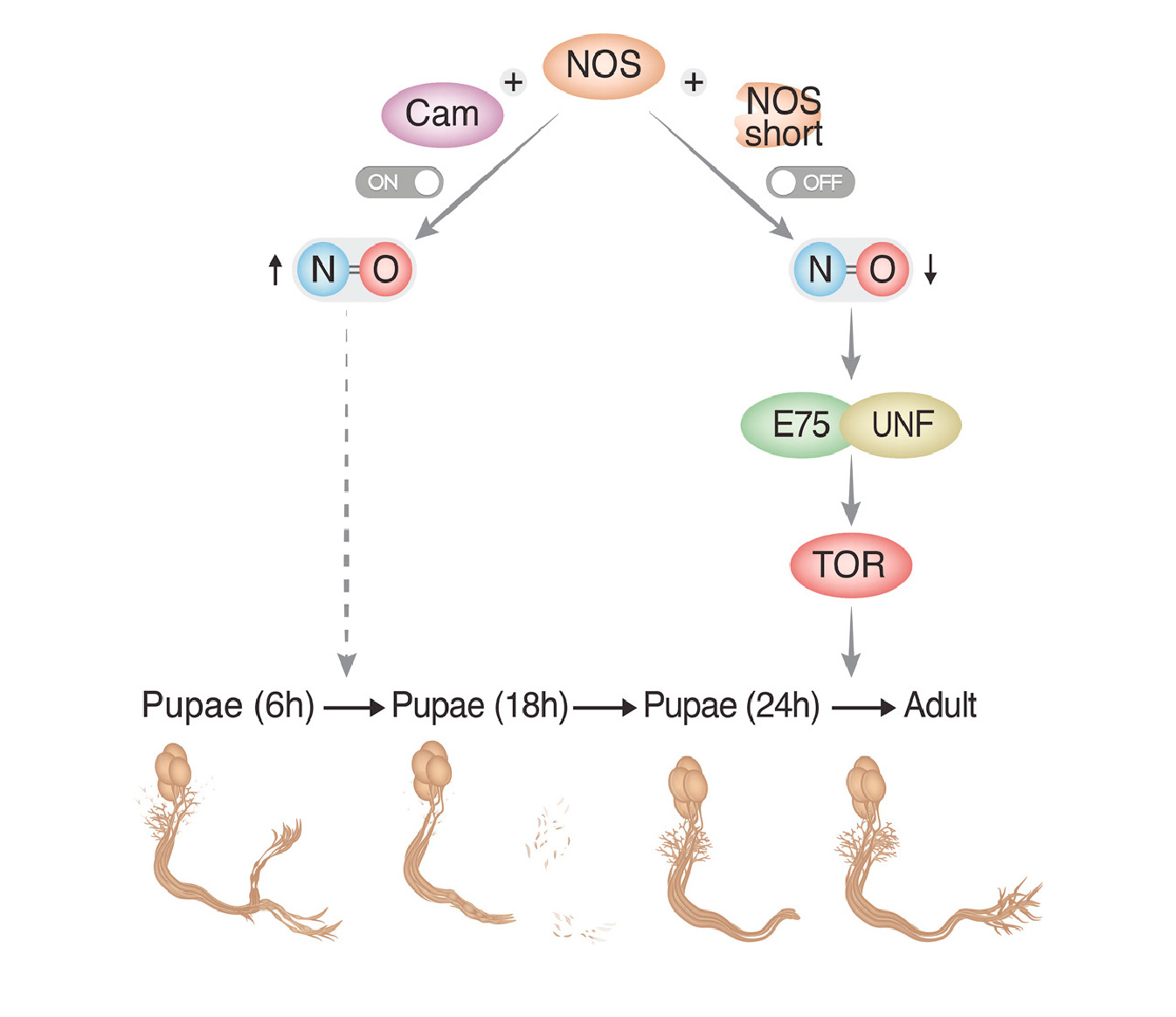
How the brain wires itself during development remains one of neurobiology’s most captivating mysteries. Neuronal remodeling - the developmental reshaping and refinement of neural connections - is essential for sculpting the mature nervous system. When this process is disrupted, it contributes to a wide range of neuropsychiatric conditions including autism, schizophrenia and Alzheimer’s disease.
In our lab, we harness the unmatched genetic power of Drosophila to dissect the mechanisms that guide the wiring and remodeling of developing neural circuits - at the molecular, cellular and functional levels. Since remodeling involves both destructive and constructive processes, our research offers a unique window into the principles underlying degenerative and regenerative events in both physiological and pathological contexts.