The atomic-resolution structures of the relatively stable (T and R) end states of several allosteric proteins are known but the pathways by which they interconvert are generally not known. We addressed this issue using GroEL as a model system by employing linear free energy relationships of physical organic chemistry such as phi-value analysis (Yifrach and Horovitz, 1998) ; Horovitz et al, 2002), double-mutant cycles (Horovitz, 1996) and correlated mutation analysis (Kass and Horovitz, 2002; Noivirt et al., 2005).
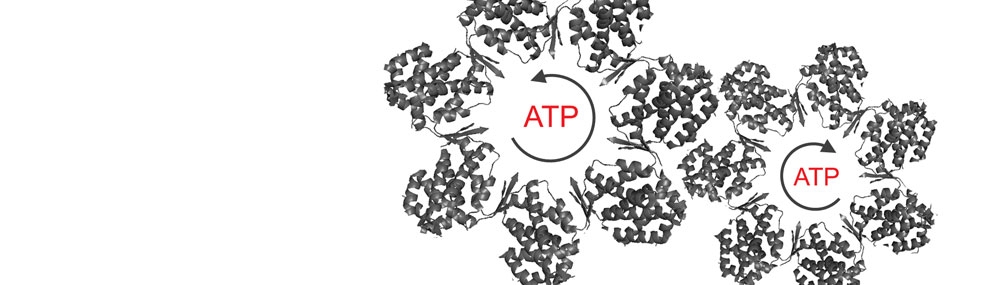
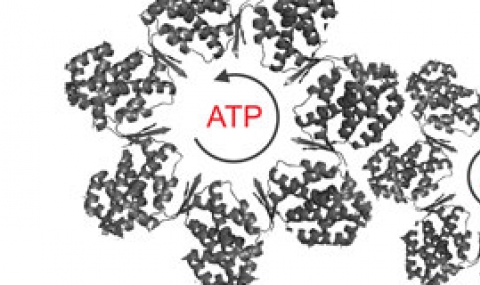
Recently, we used structural mass-spectrometry to determine the ATP-loading pathway of GroEL and show that its allosteric transitions are concerted (Dyachenko et al., 2013). In future work, we intend to apply the structural mass-spectrometry approach to other systems such as CCT/TRiC.