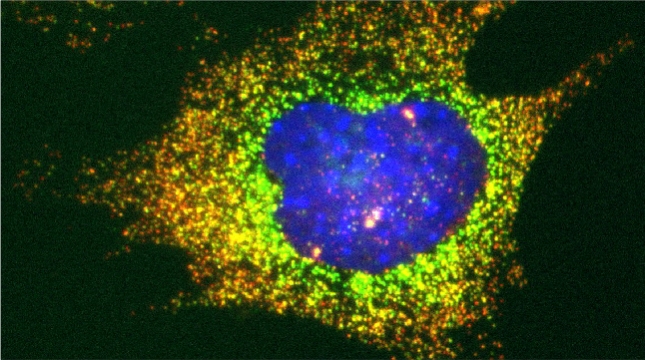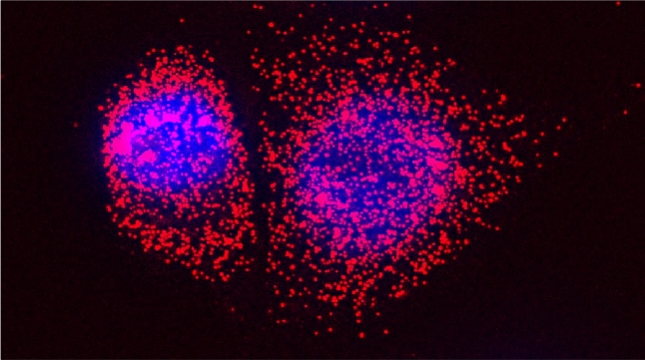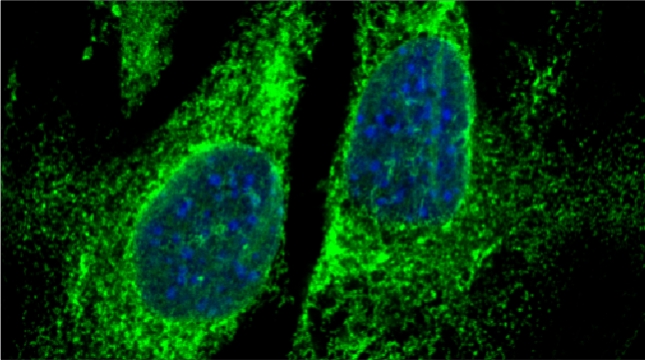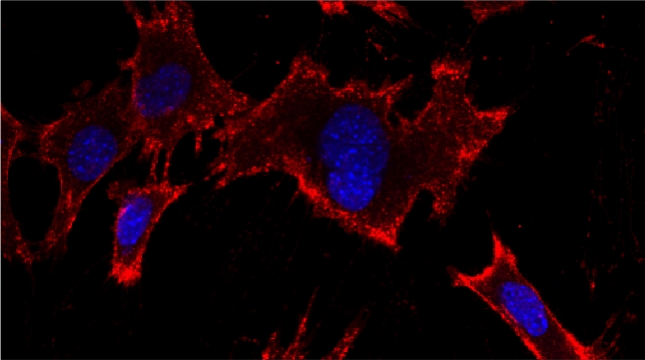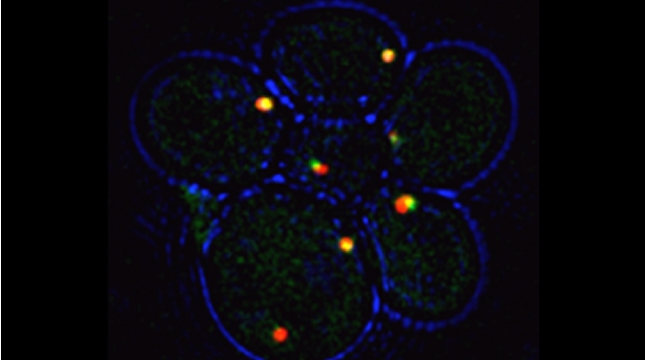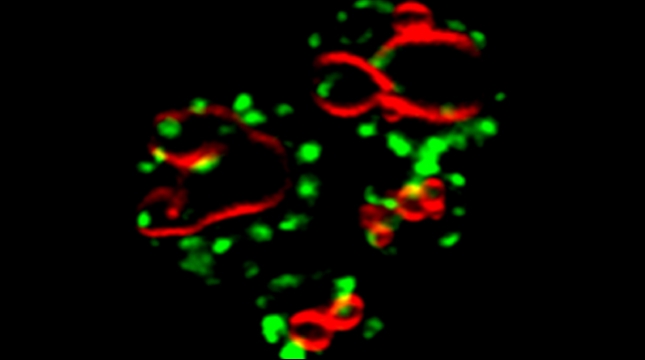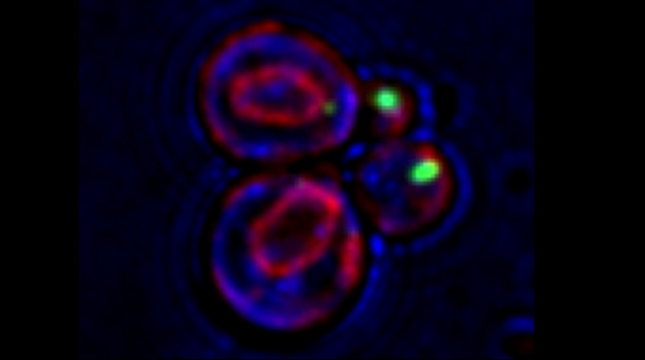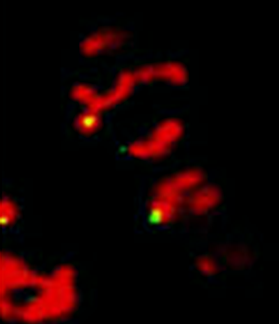
“The control of protein import into mitochondria is essential for their functions (e.g. ATP synthesis, lipid metabolism, calcium storage, etc.), however, the precise mechanism of protein import – whether it is post-translational, co-translational, or a combination thereof - is still open for debate. Yeast have >600 nuclear genes encoding for mitochondrial proteins and we are studying the process by which mRNAs encoding mitochondrial proteins (mMPs) may reach the mitochondria. By employing a novel endogenous mRNA tagging and visualization procedure developed in our lab, called m-TAG [Haim et al 2007, Haim-Vilmovsky and Gerst 2009, Haim-Vilmovsky and Gerst 2011], we have found that a number of mMPs associate with mitochondria and in a manner that is dependent upon cis-acting elements in the messages and trans-acting RNA-binding proteins [Gadir et al., 2011]. Ongoing work using RaPID, an RNA purification procedure developed in our laboratory to affinity purify aptamer-tagged mRNAs [Slobodin and Gerst, 2010 and 2011], has identified components of the COPI intracellular vesicular trafficking pathway as mediating mMP localization [Zabezhinsky et al., 2016]. Upon inactivation of this pathway, defects in mitochondrial morphology, protein import, and mitochondrial function occur and lead to cell death.