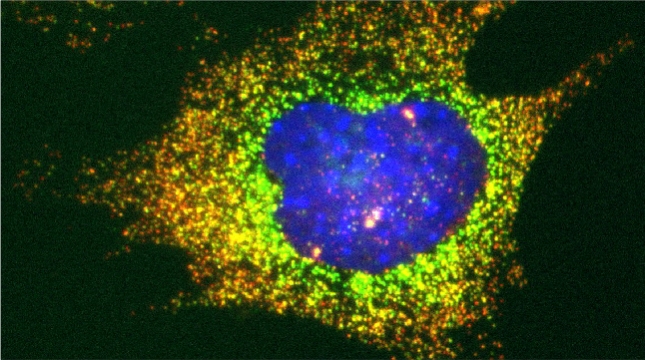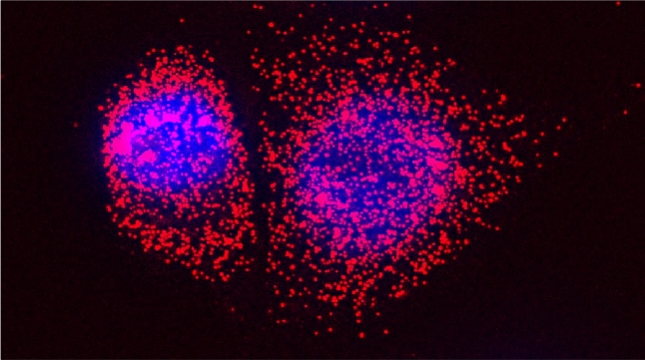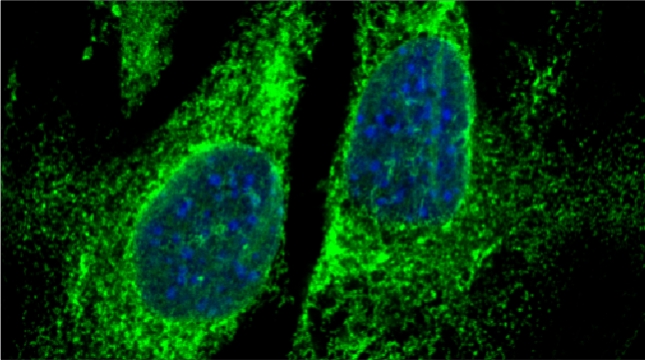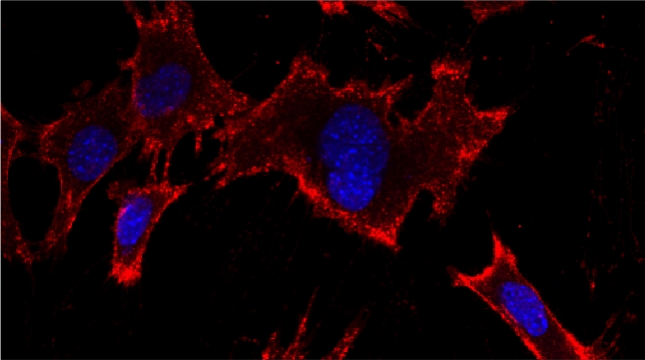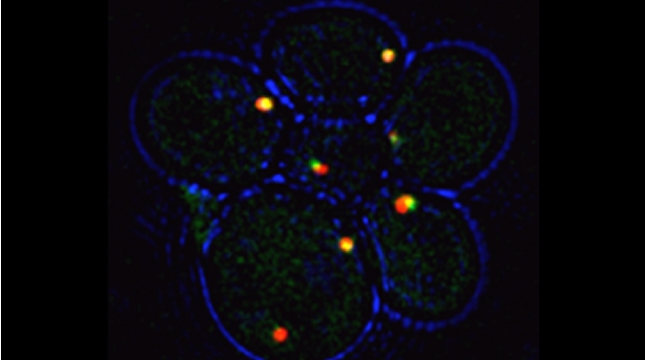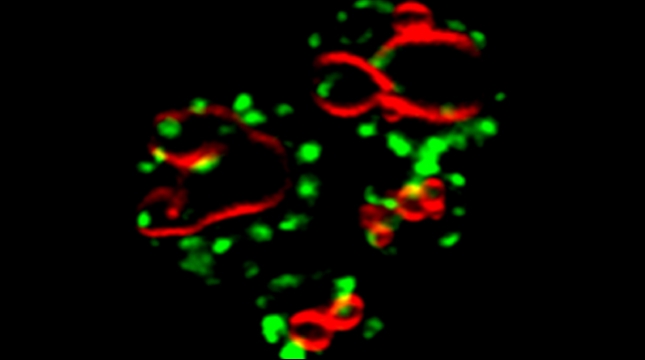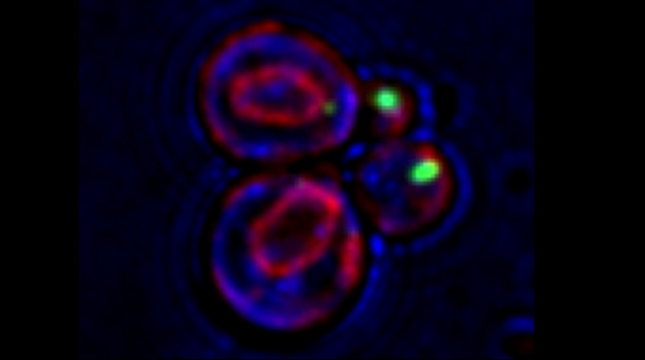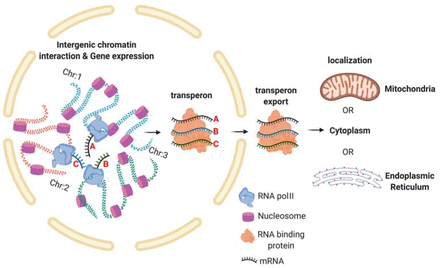
Prokaryotes utilize polycistronic messages (operons) to co-translate proteins involved in the same biological process. Whether eukaryotes achieve similar regulation by selectively assembling monocistronic messages derived from different chromosomes is unclear. We identified mRNAs that co-precipitate into ribonucleoprotein (RNP) particles in yeast. Consistent with the hypothesis of eukaryotic RNA operons, mRNAs encoding components of the mating pathway, heat shock proteins, and mitochondrial outer membrane proteins multiplex in trans to form discrete mRNP particles, termed transperons. Chromatin-capture experiments reveal that genes encoding multiplexed mRNAs physically interact, thus, RNA assembly may result from co-regulated gene expression. Transperon assembly and function depends upon H4 histones and their depletion leads to defects in RNA multiplexing, resulting in decreased pheromone responsiveness and mating, and increased heat shock sensitivity. We propose that intergenic associations and non-canonical H4 histone functions contribute to transperon formation in eukaryotic cells to regulate cell physiology (Rohini Nair et al., 2020, eLife). This mechanism seems to be conserved from yeast to mammals: we demonstrated that mRNAs encoding proteins of the heat-shock response in mouse cells also form transperons (Pataki & Gerst, 2025, Sci Adv).
We are currently looking into the molecular mechanism of mRNA multiplexing in yeast, as well as in mammalian cells.