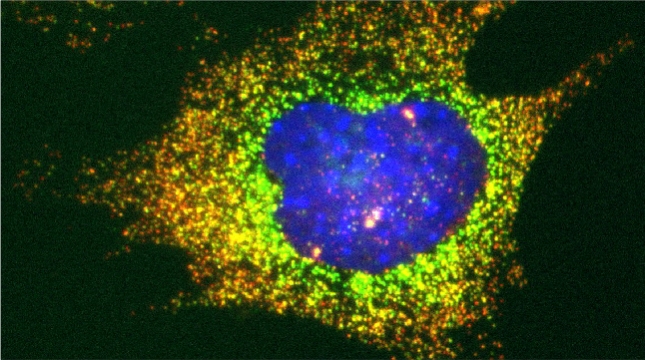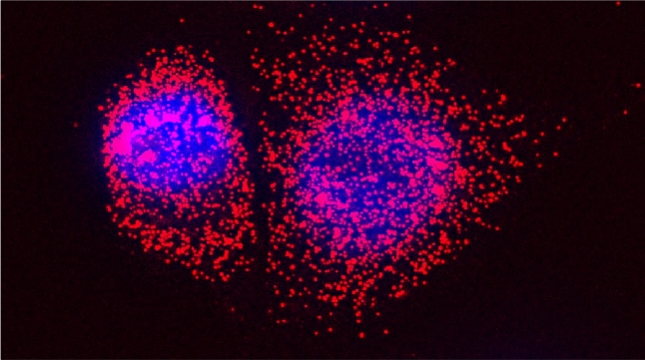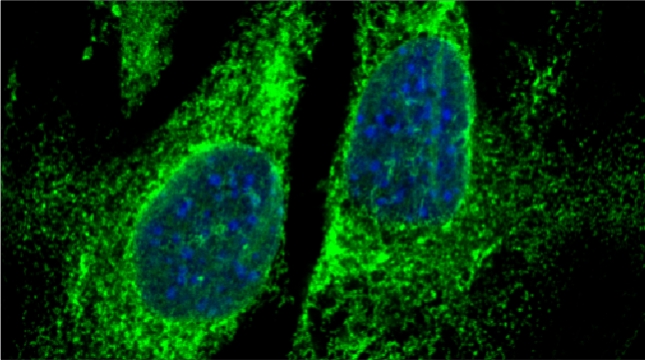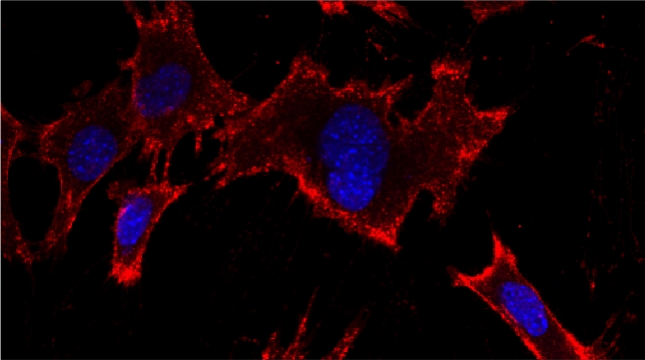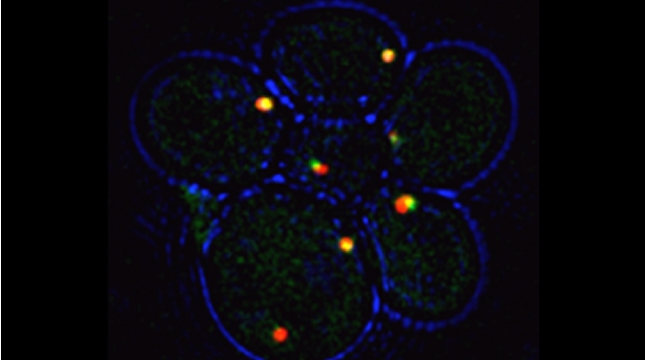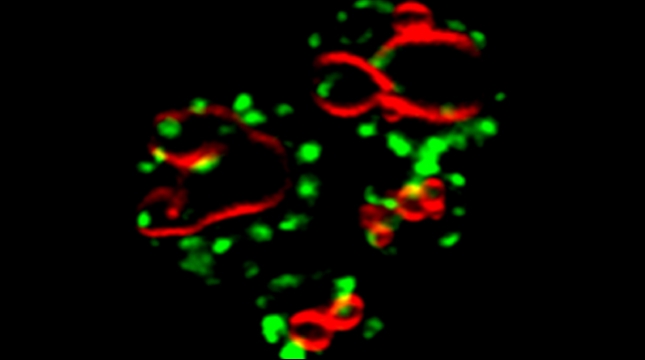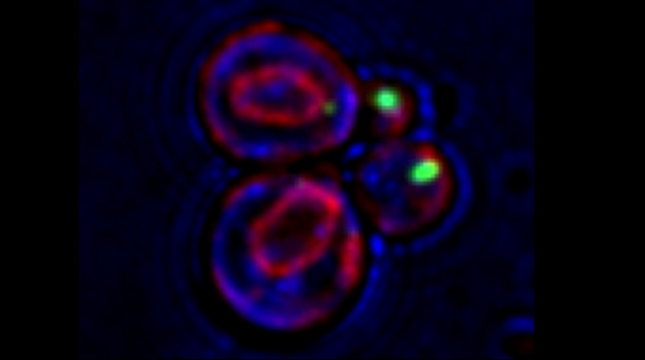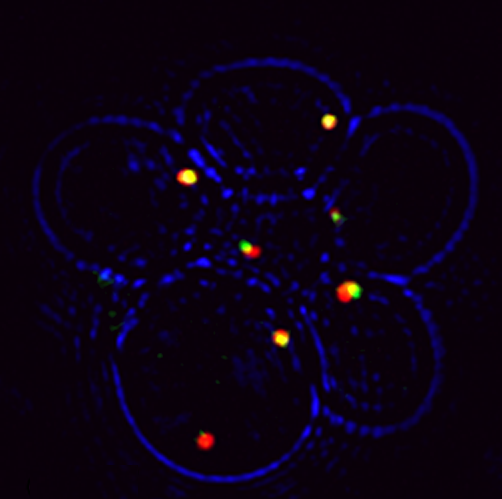
Peroxisomes play an important role in the oxidation of fatty acids, as well as in the synthesis of cholesterol and bile acids, and over 50 genes are associated with peroxisome biogenesis and function. We used our endogenous mRNA tagging procedure, m-TAG [Haim et al 2007, Haim-Vilmovsky and Gerst, 2009] to localize mRNAs encoding peroxisomal proteins (mPPs) for the first time. We found that a subset of mPPs localized at high levels to peroxisomes, while another subset only showed low levels of localization, and one mPP localized solely to the ER [Zipor et al 2009]. While the mechanism of how proteins are targeted to peroxisomes is not fully understood, there is direct evidence of post-translational import although co-translational import and endomembrane trafficking are very likely to contribute to this process. mPP targeting may require an RNA-binding protein called Puf5, although the deletion of PUF5 does not inhibit the ability of cells to grow on fatty acids, such as oleate, as the sole carbon source. Thus, other RNA-binding proteins and the cis–acting elements they interact with are likely to play a role in mPP localization. Ongoing work seeks to identify these factors that regulate mPP localization to peroxisomes and/or the ER [Haimovich et al. 2015].