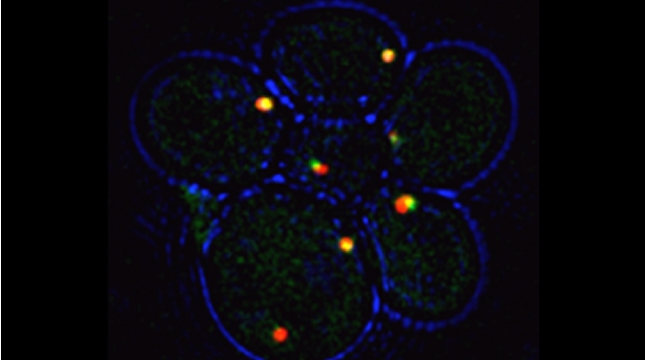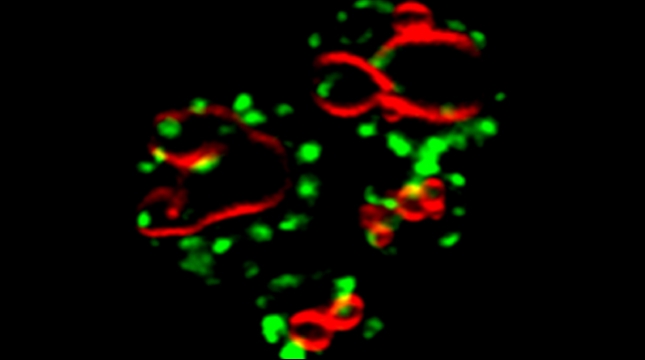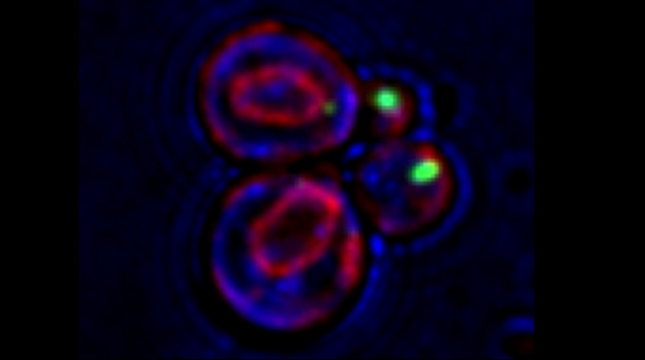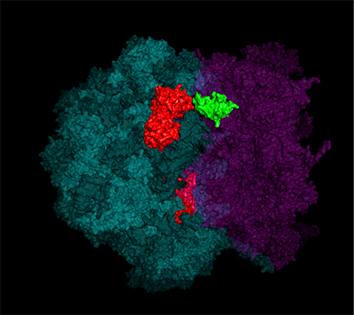
Yeast have ~140 nuclear-encoded ribosomal proteins, of which almost all are constituents of paralog pairs. Yet these paralogs have non-identical functions, whereby deletions in individual paralogs yield differences in phenotype and transcriptional profiling. One example in yeast has already demonstrated that specific paralogs are involved in the delivery and translation of ASH1 mRNA, which encodes a cell-fate component, in newly forming daughter cells.
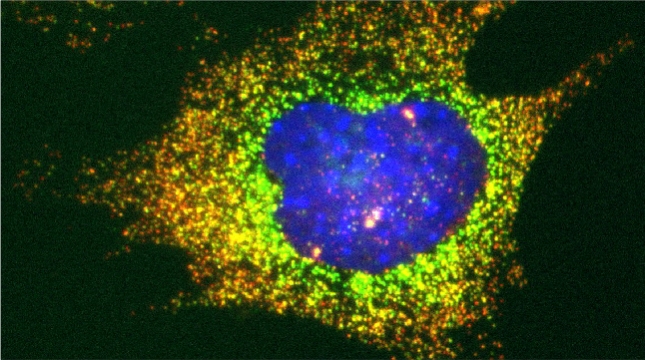
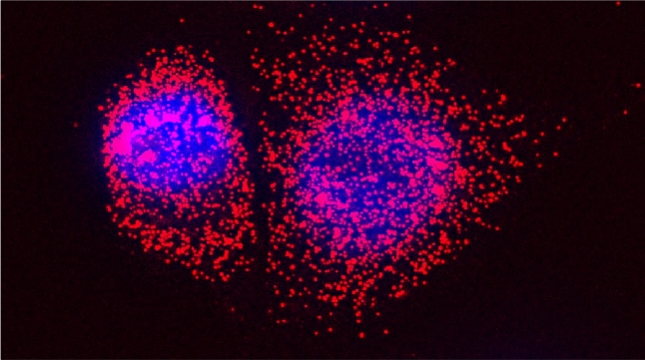
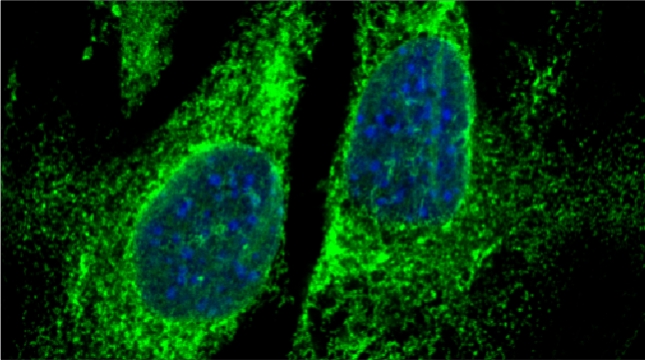
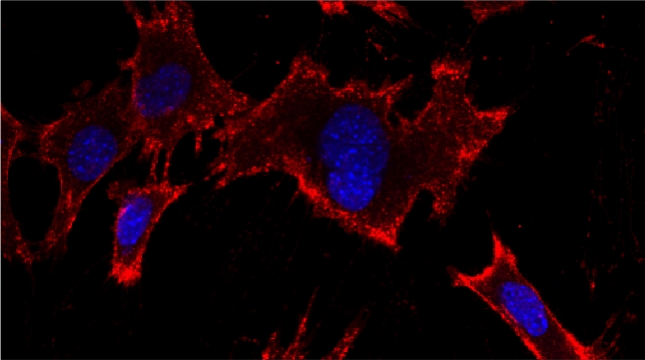
In our lab we are studying ribosomal protein paralogs involved in mitochondrial function leading to respiration and growth on non-fermentable carbon sources. Since mRNAs encoding mitochondrial proteins (mMPs) are targeted to mitochondria [Gadir et al 2011, Zabezhinsky et al., 2016], it may be that specialized cytosolic ribosomes (i.e. composed of discrete ribosomal protein paralogs) engage these mRNAs and confer their specific translation. Our work showed that specific ribosomal proteins control mMP translation and have important consequences upon mitochondrial morphology and function [Segev & Gerst 2018]. If specialized ribosomes act at the level of mitochondria, then additional ribosomal protein paralogs might act in a combinatorial fashion to regulate the translation of other subpopulations of RNAs in cells. Paralog specificity may prove to be an important means to control the cellular translatome under different growth and environmental conditions [Gerst 2018].
Indeed, we employed yeast ribosomal protein (RP) paralog deletion libraries and high-throughput screening to investigate the functional specificity and redundancy between paralogs under various growth conditions. Composition and translatome analyses verified paralog specificity in the assembly and function of ribosomes specialized for growth on different carbon sources, and identified a novel RAP required for the efficient translation of peroxisomal proteins. Importantly, we also show that the mechanism by which specific RP paralogs incorporate into ribosomes requires their unique 3’-untranslated regions to yield ribosomes that differ in composition and function [Singh et al. 2024].