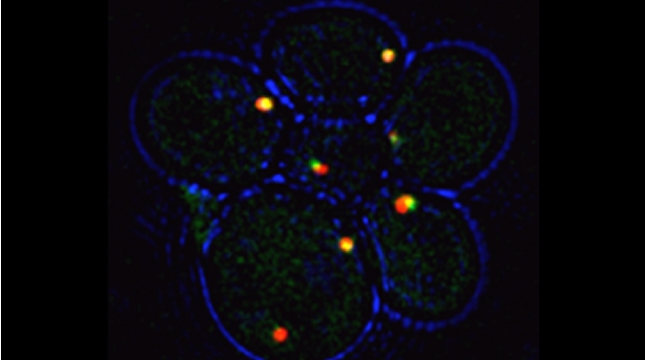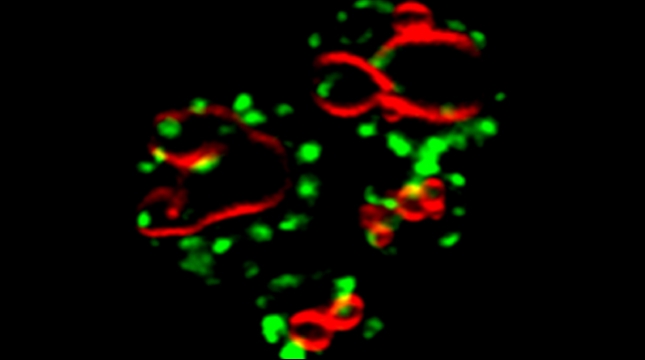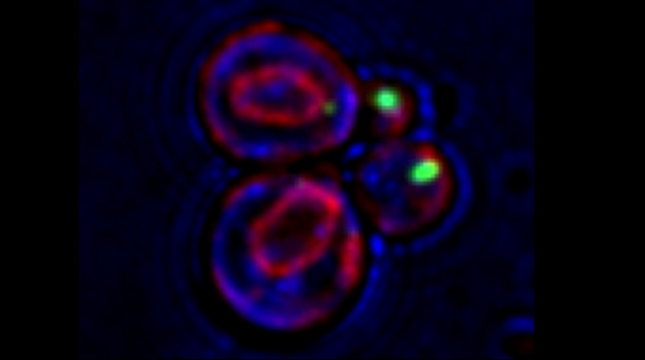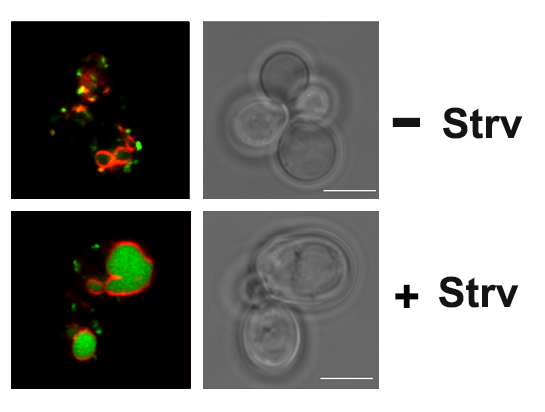
In the yeast model for Batten disease, specific trans Golgi membrane proteins re-localize to the endolysosomal pathway and undergo degradation in cells lacking BTN1, perhaps through defects in Golgi SNARE assembly and protein retrieval [Kama et al 2011]. We now find that the btn1Δ genotype phenocopies nutrient-limiting conditions, whereby functional Golgi proteins are recognized as Golgi quality control (GQC) substrates and redistribute to the vacuole in wild-type cells upon the turn-off of the TOR kinase signaling [Dobzinski et al 2015]. This process is ubiquitin- and multivesicular body (MVB) pathway-dependent, and we have identified the defective-for-SREBP-cleavage (DSC) complex as conferring this new type of protein quality control. Thus, TOR signaling controls DSC complex-mediated protein quality control via regulation of the MVB pathway, and may be implicated in Batten disease. This might suggest that neurons lacking CLN3 (the mammalian ortholog of Btn1) function may falsely perceive themselves to be in a state of nutrient deprivation.