Prof. Michal Sharon
Phone: +972-8-934-3947
Email: michal.sharon@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 157
Go to Website
We aim to discover the mechanisms that control and coordinate the activity of molecular machines involved in the protein degradation pathway. To do so we apply novel native mass spectrometry approaches, in conjunction with fluorescence microscopy, biochemical and cell biology methods - generating an integrative mode of analysis combining in vitro and in vivo findings.
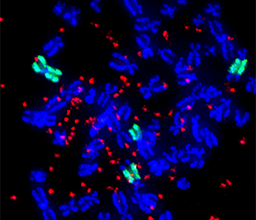

Dr. Ofer Shoshani
Phone: +972-8-934-2443
Email: ofer.shoshani@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 207, 217-219
Go to Website

We focus on studying the mechanisms that promote chromosome instability, a major driver of cancer initiation, metastasis, and drug resistance. Our group aims to understand the plasticity of the cancer genome and how it contributes to disease progression. To do so, we apply chromosome manipulation approaches in cells, to study how single genomic perturbations affect population trajectories. Our goal is to identify vulnerabilities in mechanisms driving chromosome evolution, in order to design next-generation therapies with reduced resistance in patients.

Prof. Ruth Scherz-Shouval
Phone: +972-8-934-2299
Email: ruth.shouval@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 275
Go to Website
For tumors to expand, metastasize, and evade immune surveillance, genetically transformed cancer cells must recruit non-malignant cells, including macrophages, fibroblasts and endothelial cells. These cells, collectively termed the tumor microenvironment, are reprogrammed to support the tumor at the expense of its host. Our group aims to elucidate the mechanisms by which tumors reprogram their local environments. Our goal is to provide a deeper understanding of how tumors develop into systemic malignancies, predict which tumors are more likely to do so, and design therapeutic strategies to overcome these malignancies by targeting genetically stable elements in the tumor microenvironment.


Prof. Avraham Yaron
Phone: +972-8-934-6240
Email: avraham.yaron@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 417
Go to Website

We are investigating the mechanisms that govern the wiring of the nervous system during development, which includes axonal guidance, pruning and the role of local protein synthesis.

a research lab
to join?
interactive movie
to help you choose
Looking for a research lab to join?
The department of Biomolecular Sciences proudly presents an interactive movie that will help you decide which lab you would like to join for your research work. Press on the questions that interest you to begin your journey. You can always go back or and choose another path or start over.
Navigation buttons are on the bottom and you can step back using the arrow on the upper left corner.


Prof. Michael Fainzilber
Phone: +972-8-934-4266
Fax: +972-8-934-4112
Email: mike.fainzilber@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 405
Go to Website

We are investigating long distance signaling mechanisms in neurons and other large eukaryotic cells, primarily an importins/molecular motor-based mechanism that integrates cytoplasmic and nuclear signaling. Currently our main focus is on the roles of this and related mechanisms in neuronal responses to injury and in cellular length sensing.


Prof. Rivka Dikstein
Phone: +972-8-934-2117
Email: rivka.dikstein@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 329
Go to Website
We are studying regulation of transcription and translation in health and disease. Specifically we elucidate how the transcription and translation processes control the cellular response to enviromental stimuli, we investigate the connections between the transcription and translation processes and we develop pharmacological tools to manipulate these processes.


Prof. Sarel-Jacob Fleishman
Phone: +972-8-934-6361
Email: sarel.fleishman@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 177
Go to Website

We are interested in how protein function is encoded in the structures of protein binders, enzymes, and antibodies. To test our understanding we computationally design new protein functions not seen in Nature and experimentally characterize these designs. Iterations of design and experimental characterization enable us to understand new features of how protein function is specified in Nature.


Prof. Gad Asher
Phone: +972-8-934-6949
Fax: +972-8-934-6010
Email: gad.asher@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 277
Go to Website

Our main research interest is to explore the interplay between the cellular metabolic state and circadian clocks and further identify the underlying molecular mechanisms.


Dr. Nir Fluman
Phone: +972-8-934-6456
Email: nir.fluman@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 305
Go to Website

Proteins mediate nearly all of the essential molecular processes of life. To perform their function, they need to fold into unique structures. At any given moment, thousands of proteins are produced and need to fold in the cell. But folding is an incredibly complex process that very often fails. To deal with this challenge, the cell is equipped with an array of chaperones and proteases, performing quality control functions to safeguard the cell. Failure of these processes underlies numerous devastating diseases. We investigate how these events unfold for membrane proteins, which are proteins that reside at the membrane and make up a quarter of the proteome in every living organism.


Dr. Tslil Ast
Phone: +972-8-934-6226
Email: tslil.ast@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 473
Go to Website

Mitochondria are pivotal to the function of eukaryotic cells- WHY? It’s becoming clear that one of their most fundamental and vital roles involves the biosynthesis of iron-sulfur (Fe-S) clusters. These ancient cofactors are crucial for proteins engaged in DNA replication, translation, metabolism, and cellular respiration and more. Given these critical functions, it is not surprising that disruptions in Fe-S cluster synthesis are associated with various human diseases, including the most prevalent mitochondrial disorder (Friedreich’s ataxia). Despite its importance, much remains unknown about this pathway, particularly how it is regulated within the cell. Our lab utilizes cutting-edge genetic, cell biology, and biochemical techniques to unravel this vital pathway and integrate it within the broader context of human cellular function.


Prof. Rivka Dikstein
Phone: +972-8-934-2117
Email: rivka.dikstein@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 329
Go to Website

We are studying regulation of transcription and translation in health and disease. Specifically we elucidate how the transcription and translation processes control the cellular response to enviromental stimuli, we investigate the connections between the transcription and translation processes and we develop pharmacological tools to manipulate these processes.


Dr. Ofer Shoshani
Phone: +972-8-934-2443
Email: ofer.shoshani@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 207, 217-219
Go to Website

We focus on studying the mechanisms that promote chromosome instability, a major driver of cancer initiation, metastasis, and drug resistance. Our group aims to understand the plasticity of the cancer genome and how it contributes to disease progression. To do so, we apply chromosome manipulation approaches in cells, to study how single genomic perturbations affect population trajectories. Our goal is to identify vulnerabilities in mechanisms driving chromosome evolution, in order to design next-generation therapies with reduced resistance in patients.

a research lab
to join?
interactive movie
to help you choose
Looking for a research lab to join?
The department of Biomolecular Sciences proudly presents an interactive movie that will help you decide which lab you would like to join for your research work. Press on the questions that interest you to begin your journey. You can always go back or and choose another path or start over.
Navigation buttons are on the bottom and you can step back using the arrow on the upper left corner.


Prof. Sarel-Jacob Fleishman
Phone: +972-8-934-6361
Email: sarel.fleishman@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 177
Go to Website

We are interested in how protein function is encoded in the structures of protein binders, enzymes, and antibodies. To test our understanding we computationally design new protein functions not seen in Nature and experimentally characterize these designs. Iterations of design and experimental characterization enable us to understand new features of how protein function is specified in Nature.


Prof. Ruth Scherz-Shouval
Phone: +972-8-934-2299
Email: ruth.shouval@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 275
Go to Website

For tumors to expand, metastasize, and evade immune surveillance, genetically transformed cancer cells must recruit non-malignant cells, including macrophages, fibroblasts and endothelial cells. These cells, collectively termed the tumor microenvironment, are reprogrammed to support the tumor at the expense of its host. Our group aims to elucidate the mechanisms by which tumors reprogram their local environments. Our goal is to provide a deeper understanding of how tumors develop into systemic malignancies, predict which tumors are more likely to do so, and design therapeutic strategies to overcome these malignancies by targeting genetically stable elements in the tumor microenvironment.

Prof. Neta Regev-Rudzki
Phone: +972-8-934-3160
Email: neta.regev-rudzki@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 205
Go to Website
With malaria continuing to be a major global disease, advances toward understanding the basic biology of P. falciparum remain essential. Our studies focus on different aspects of the cellular biology of the malaria parasite. In particular, we aim to explore cell-cell communication pathways between the parasites themselves and their human host.


Prof. Ziv Reich
Phone: +972-8-934-2982
Email: ziv.reich@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 259
Go to Website

We are interested in transport processes and in photosynthesis. Within the realm of photosynthesis we are mainly concerned with dynamic processes that accompany the life cycle of the thylakoid network, including its response to different stresses and its formation and dismantling. Regarding nucleo-cytoplasmic transport, we are particularly interested in its selectivity, the behavior of the ensemble of transporting molecules as it relates to the transport of a single molecule and in applications to gene therapy. In both fields of study, we combine different approaches and methodologies including ensemble and single-molecule biophysical methods, biochemical and molecular biology techniques, statistical mechanical modeling and state-of-the-art electron microscopy.
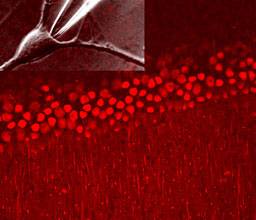

Prof. Eitan Reuveny
Phone: +972-8-934-3243
Fax: +972-8-934-2135
Email: e.reuveny@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 356
Go to Website
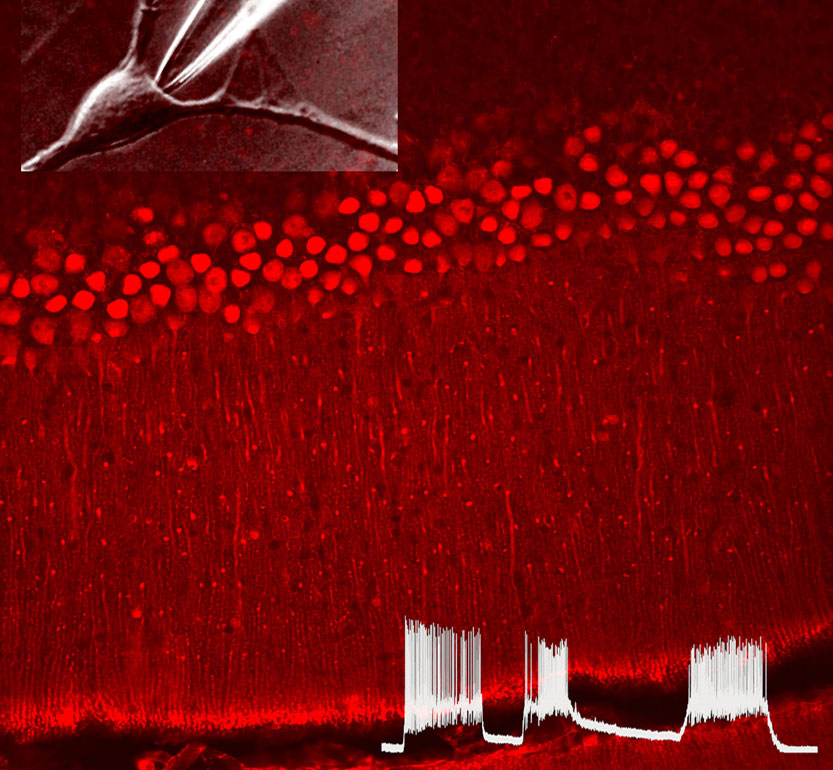
We are interested in deciphering the mechanisms by which extracellular and intracellular signaling queues affect ion channel function, both in excitable and non-excitable tissues. We focus on three main areas: 1) GPCRs-mediated ion channel regulation and modulation, 2) cellular mechanisms that control calcium homeostasis, 3) how physiological functions are affected by ions channels activity.

Prof. Zvi Livneh
Phone: +972-8-934-3203
Fax: +972-8-934-4169
Email: zvi.livneh@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 307
Go to Website

We are studying molecular mechanisms of DNA repair, and in particular error-prone repair (translesion DNA synthesis) in mammalian cells. In addition, we employ the knowledge on DNA repair to develop novel functional enzymatic DNA repair biomarkers for risk assessment and prevention of cancer.


Prof. Ziv Reich
Phone: +972-8-934-2982
Email: ziv.reich@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 259
Go to Website

We are interested in transport processes and in photosynthesis. Within the realm of photosynthesis we are mainly concerned with dynamic processes that accompany the life cycle of the thylakoid network, including its response to different stresses and its formation and dismantling. Regarding nucleo-cytoplasmic transport, we are particularly interested in its selectivity, the behavior of the ensemble of transporting molecules as it relates to the transport of a single molecule and in applications to gene therapy. In both fields of study, we combine different approaches and methodologies including ensemble and single-molecule biophysical methods, biochemical and molecular biology techniques, statistical mechanical modeling and state-of-the-art electron microscopy.


Dr. Ori Avinoam
Phone: +972-8-934-3557
Email: ori.avinoam@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 374
Go to Website
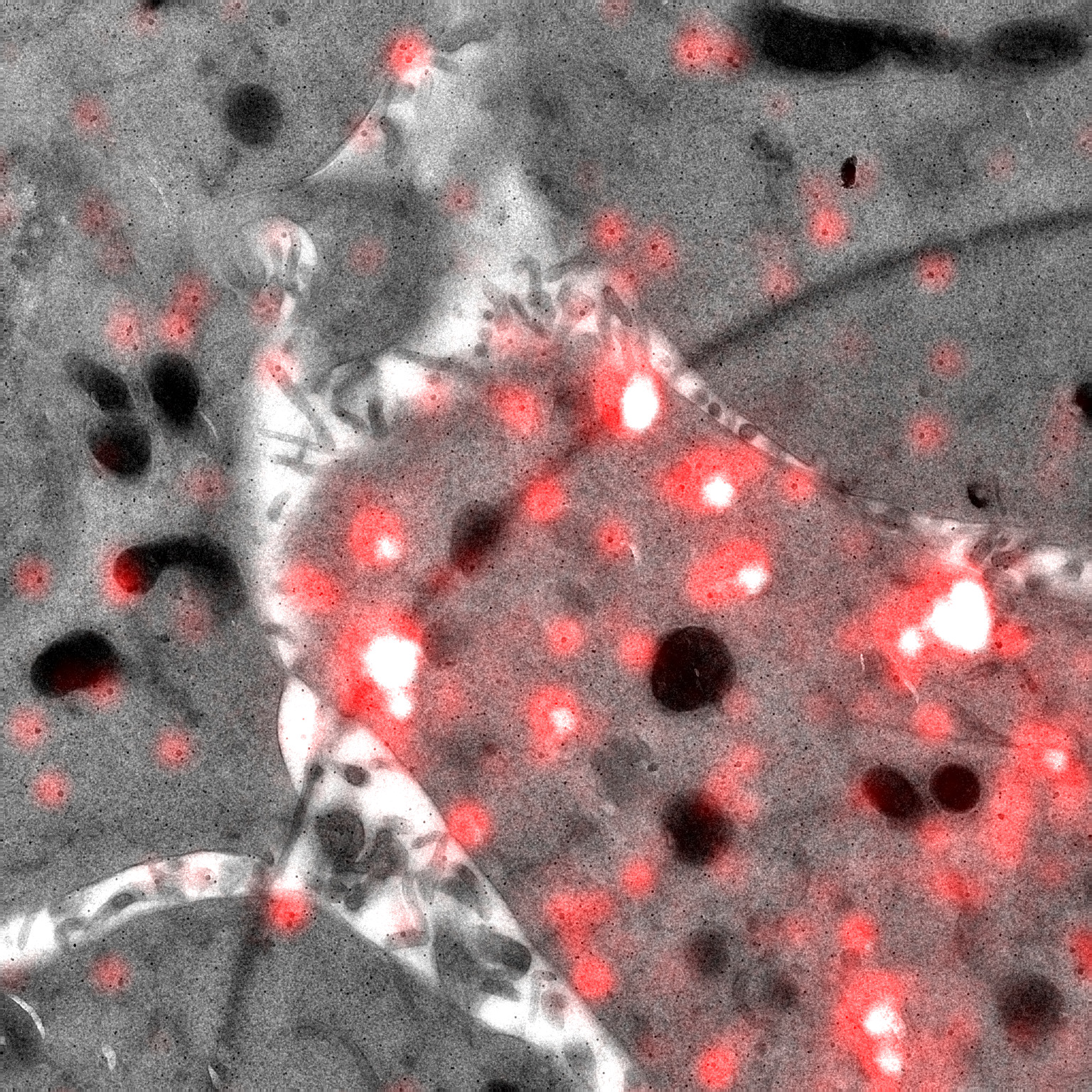
We are interested in the universal process of membrane remodeling that occurs during endocytosis, when subdomains of the membrane are reshaped into cargo transport vesicles, which allow cells to take up material and communicate with the environment. Our studies focus on understanding the molecular mechanisms of endocytosis and their contribution to cell physiology. We use a combination of advanced imaging techniques that include correlated light and electron microscopy to visualize these processes with high spatial and temporal resolution. We are particularly interested in understanding how endocytic processes contribute to muscle differentiation and homeostasis.
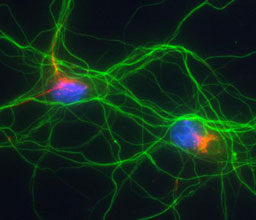

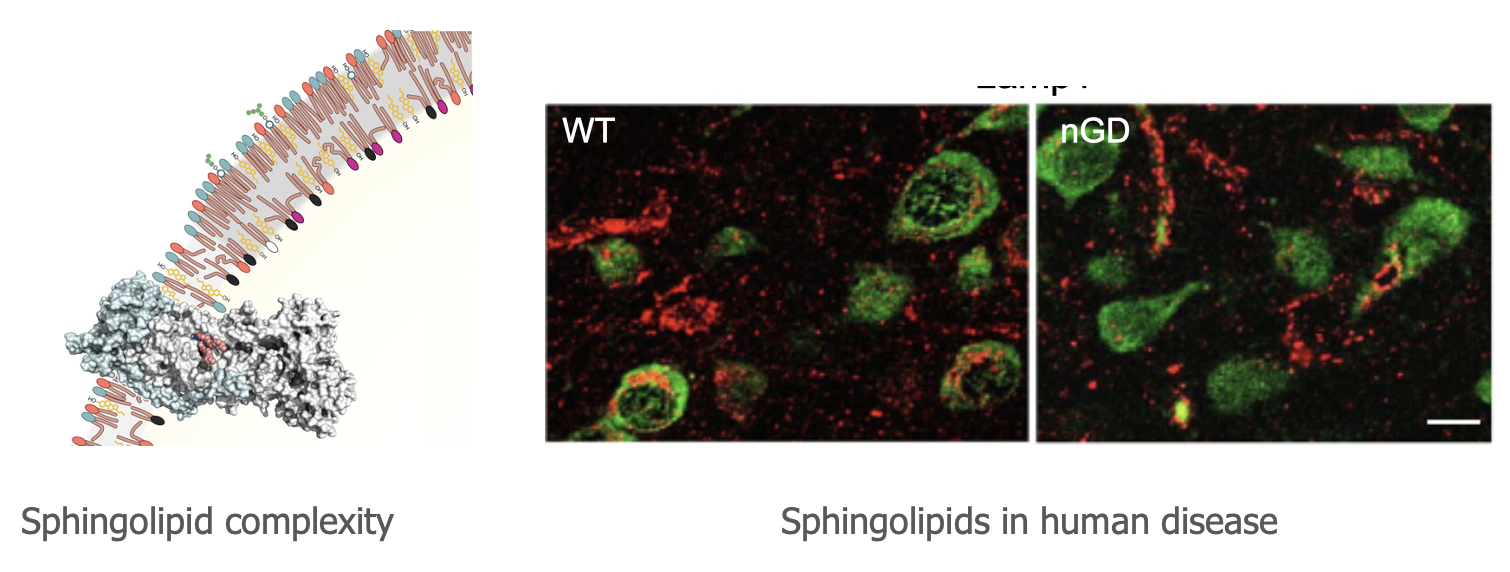
Prof. Anthony H. Futerman
Phone: +972-8-934-2704
Fax: +972-8-934-4112
Email: tony.futerman@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 456
Go to Website


Prof. Gideon Schreiber
Phone: +972-8-934-3249
Email: gideon.schreiber@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 475
Go to Website
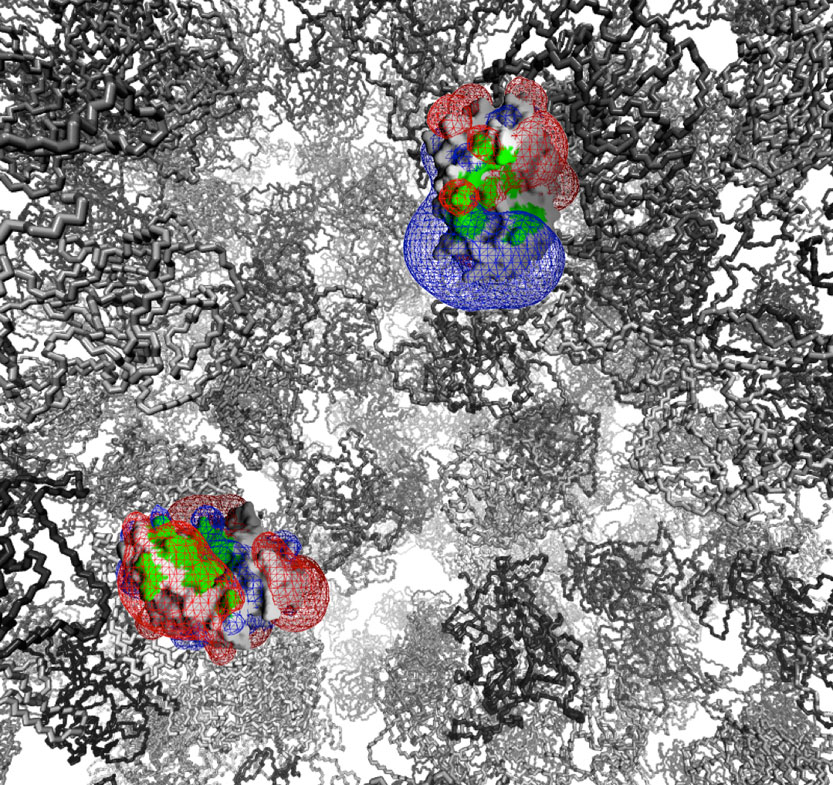
We study the basic physico-chemical principles governing the kinetics, thermodynamics and specificity of protein-protein interactions. The gained knowledge is translated into the development and implementation of protein engineering methodologies. Between other, these are used to study how the interaction of interferon with its receptors results in complex differential responses in cells.


Prof. Neta Regev-Rudzki
Phone: +972-8-934-3160
Email: neta.regev-rudzki@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 205
Go to Website

With malaria continuing to be a major global disease, advances toward understanding the basic biology of P. falciparum remain essential. Our studies focus on different aspects of the cellular biology of the malaria parasite. In particular, we aim to explore cell-cell communication pathways between the parasites themselves and their human host.
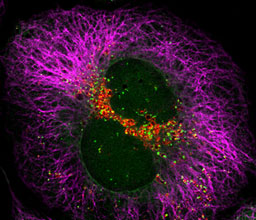

Prof. Zvulun Elazar
Phone: +972-8-934-3682
Email: zvulun.elazar@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 429
Go to Website

We study the molecular mechanism and regulation of autophagy, a process responsible for the degradation of cytoplasmic proteins and organelles. A particular emphasis is given to the relationship of this catabolic pathway to disease such as cancer and neurodegeneration.


Prof. Yoav Soen
Phone: +972-8-934-6011
Email: yoavs@weizmann.ac.il
Location: Nella and Leon Benoziyo Building for Biological Sciences, Room 229
Go to Website

We study epigenetic mechanisms by which developmental plasticity allows the environment to bring about heritable modifications in the developmental program. We approach this with a combination of in vitro (mammalian cells) and in vivo models (the fly, D. melanogaster), in which we confront developmental processes with unforeseen environmental challenges that promote deviations from the selected patterns of development.