
Yaron Emanuel Antebi
Signal processing in cellular decision making
- Developing a systems-level framework to characterize signal integration at the single cell level
- Defining the role of perception and integration of complex stimuli in determining cellular fate
- Analyzing multi-ligand information processing in the BMP/TGFβ pathway and its effect on mesenchymal stem cell differentiation
- Studying signal processing in the JAK/STAT pathway during hematopoiesis and immune response
room 415A


Eli Arama
Programmed Cell Death and Cellular Destruction Mechanisms in Development
- Caspase-dependent non-lethal cellular processes (CDPs) in Drosophila:
- Sperm terminal differentiation
- Inhibition of unwanted cell migration and invasion
- Caspase-independent alternative cell death pathways (ACDs) in Drosophila development
- Germ Cell Death (GCD) during premeiotic germ cell formation in the adult males
- Primordial Germ Cell (PGC) death during germ cell specification in the embryo
- Structural organization of the Drosophila sperm mitochondria (Nebenkern)
- Paternal Mitochondrial Destruction after fertilization in Drosophila

Naama Barkai
Principles of Biological Circuits
- Biological circuits are the building blocks of cellular information processing: what are the principles guiding their design, function and evolution?
- Basic biological processes such as gene expression rely on a small number and therefore vary in a stochastic, unpredictable way. What is the role of variability in information processing and what mechanisms ensure robustness of processes such as embryonic patterning?
- Transcription and replication compete for the same DNA template, and rely on the combined action of general and specific regulatory factors. How are these processes coordinate and how do they communicate with each other?

Ari Elson
Cell signaling in osteoclasts: bone in health and disease
- We study how osteoclasts, the only cells in our bodies that can degrade bone, are formed and how they function
- Our studies are directly connected to bone biology and to human disease, including osteoporosis, osteopetrosis, and bone loss in cancer
- Our studies combine between the molecular, cellular, and whole-organism (mouse model) levels
- Our goals are to explain how individual signaling molecules and processes regulate normal physiology, how they can contribute to disease, and how they can be used to treat disease


Jeffrey Gerst
Intracellular and intercellular mRNA trafficking and protein localization
- Labeling and live imaging of mRNAs in yeast and mammalian cells
- Affinity purification of mRNAs and identification of RNA-binding proteins and co-trafficked mRNAs
- Assessment of the role of RNA-binding proteins in protein localization and cell physiology
- Genome-wide mapping of mRNA localization and RNA-RNA and RNA-protein interactions


David Gokhman
Human evolution and gene regulation
- What are the genetic changes that made us human?
- What is the origin of human-specific diseases?
- How can we computationally infer traits from genetic data?
-
?What is the genetic basis of human adaptation


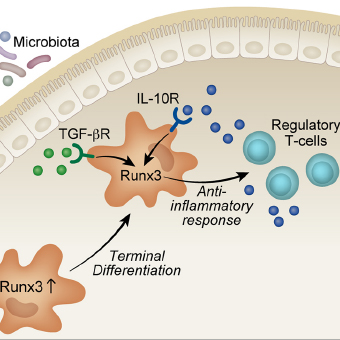
Yoram Groner
RUNX transcription factors in development and disease
- Regulatory elements conferring time and tissue specific expression of RUNX transcription factors
- Runx3 target genes in TrkC neurons
- Function of Runx3 in gut macrophages and dendritic cells
- Runx1 transcriptionally regulates megakaryocytic maturation and myoblast proliferation

Dvir Gur
Biological Crystallization Mechanisms
- Study the development and cell biology of crystal-forming cells in model organisms (zebrafish, medaka) and exotic species (Copepods, Chameleon, Geckos).
- Study pathological crystallization (i.e. gout disease and kidney stones) and develop new therapeutic approaches.
- Discover and characterize new bio-crystals.
- Investigate novel crystal-based biological optical systems.


Jacob (Yaqub) Hanna
Pluripotent Stem cells and Epigenetics
- How do mature differentiated cells revert back to an embryonic pluripotent state?
- How do different flavors of embryonic pluripotency influence differentiation potential of stem cells
- What are the key epigenetic pathways regulating timely execution of lineage commitment programs during early development?
- Why do totipotent or primordial germ cells retain only some of properties of embryonic stem cells?
room 005


Eran Hornstein
Regulatory RNA in brain integrity and neurodegeneration
- We are RNA biologists, seeking to understand RNA function in lifelong tissues: motor neurons and β cells.
- A long-term interest is on neurodegeneration, focusing on ALS. Our research spans from basic molecular mechanisms to translational / clinical studies.
- Human genomics, transcriptomics and proximity proteomics drive current projects:
(i) Specific non-coding RNAs and regulated pathways in lifelong cells
(ii) Decompensated microRNA biogenesis contribute to ALS pathogenesis
(iii) Hunting non-coding mutations, which drive ALS by human ALS genomics
(iv) Cell-free RNA biomarkers for neurodegeneration
(v) Silencing of disallowed genes in the endocrine pancreas
(vi) microRNA contribution to adult β-cell homeostasis and cellular plasticity
room 315

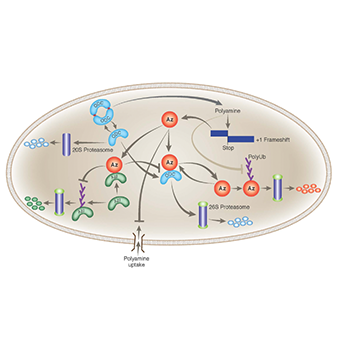
Chaim Kahana
Role of polyamines in supporting cellular functions
- Polyamines and cellular proliferation
- Polyamines in regulating translation
- Mechanism of degradation of proteins from the polyamine biosynthesis pathway

Eyal Karzbrun
Self-organization in embryonic development:
- Developing 3D human stem-cell systems as minimal quantitative models of embryonic organ formation.
- Studying the physical principles of tissue morphogenesis.
- Defining how geometry and morphogen gradients encode organ shape.
- Understanding how genetic mutations drive organ shape malformations in disease.


Adi Kimchi
The Protein Interaction Map of Programmed Cell Death in Development and Disease
- Monitoring the global profile of protein- protein interactions along the autophagic and apoptotic pathways, in cells in real time
- Whole genome functional screens to identify drivers of alternative forms of programmed cell death
- Non canonical modes of RNA translation in embryonal stem cell differentiation
- Mechanisms of cell death in the developing mammalian embryo
- Identifying the cell death signature of patient’s tumors and targeting point of vulnerability towards precision cancer therapy


Doron Lancet
Systems Biology, Medicine and Protobiology
- A Systems Biology and Systems Medicine suite of databases: GeneCards, MalaCards and PathCards
- GeneHancer: Integrated view of gene regulatory elements and their taget genes
- Systems Protobiology: Origin of Life in lipid catalytic networks without RNA
- Deciphering genetic diseases with VarElect, a whole genome sequence phenotype interpretation tool


Orly Laufman
Molecular and Cell Biology of RNA viruses
- RNA viruses including corona, zika and dengue are a major threat to human health. We study how RNA viruses interact with their host cells and transform them into viral manufactories.
- Our main model is enteroviruses - common viral pathogens that cause severe medical complications in humans, with no available therapeutic treatments.
- We investigate how enteroviruses remodel the structure and function of host organelles to generate an environment favorable to virus replication.
- Another key question we tackle is how enteroviruses, that express only a small number of proteins, take control of human cells with complex protein machineries. We study the different roles of viral proteins and the mechanisms they use to hijack host machineries.
- We aim to piece together the complete program of enterovirus replication. This could lead to the development of new antiviral therapeutics.


Shmuel Pietrokovski
Computational Biology of Conservation and Change
- Evolution, function, and activity of intein protein domains, intein-like protein-domains, and intein-associated homing endonucleases
- Sex-specific gene expression and Human genetics
- Computational genomics of predatory bacteria
- Developing methods and procedures for analysing sequence and structure of proteins, genes and genomes
room 326


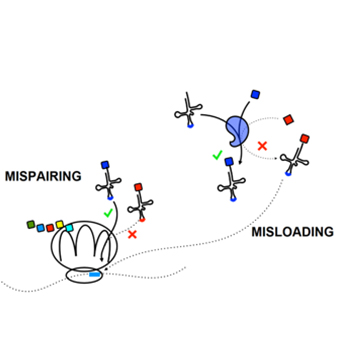
Yitzhak Pilpel
Genome Evolution and Systems Biology
- Evolution of genomes and gene expression regulatory networks
- Cancer genome, transcriptome and proteome
- Systems and genome biology of translation regulation
- Physiology of cellular response to the environment
- Evolution of human language

Orly Reiner
Cortical Development in Health and Disease
- Understanding the molecular and cellular events that shape the developing human cortex
- Studying the functions of neurodevelopmental disease-causing genes
- Developing mouse models for human neurodevelopmental disorders
- Applying “Disease in a Dish” approach for studying human embryonic cells derived brain organoids


Menachem Rubinstein
Leukotrienes link ER stress and oxidative stress
- Endoplasmic reticulum (ER) stress and cytotoxic agents trigger biosynthesis of Leukotriene C4 (LTC4) by activating a biosynthetic pathway based on microsomal glutathione S transferase 2 (MGST2)
- LTC4, in turn, triggers oxidative stress and DNA damage by nuclear translocation of NADPH oxidase 4 (NOX4)
- LTC4 inhibitors, which serve as asthma drugs alleviated cytotoxicity of chemotherapeutic agents


Maya Schuldiner
Systematic Cell Biology of Organelles
- Search for functions for unstudied proteins in yeast
- Understand how organelles function
- Study how proteins are targeted and translocated into the endoplasmic reticulum, mitochondria and peroxisomes
- Uncover new contact sites, their tethering proteins and functions
- Focus on peroxisomes, metabolic hubs of the cells
room 122

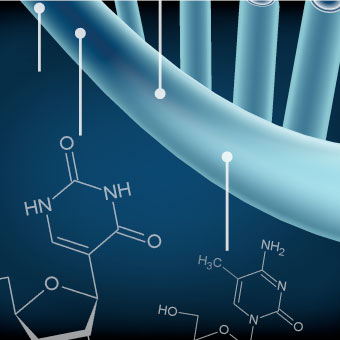
Schraga Schwartz
Cracking the epitranscriptome
- Mapping and characterizing the growing repertoire of post-transcriptional modifications present on mRNA ('the epitranscriptome')
- Deciphering the functions of post-transcriptional modifications in regulating mRNA fate (localization/stability/translation) and cellular physiology
- Dissecting the mechanisms through which mRNA modifications act
- Interdisciplinary integration of Molecular Biology, Genetics and Computational Biology

Yosef Shaul
Molecular virology and cell biology
- Understanding the molecular basis of virus-host interaction using HBV and Py viruses as models
- The role of non-receptor tyrosine kinases in DNA damage and Hippo pathways in cell fate determination
- The process of proteasomal degradation of intrinsically disordered proteins (IDPs)


Rotem Sorek
Microbial genomics and systems biology
- CRISPR, the microbial anti-phage immune system
- The arms-race between bacteria and phage
- Communication between viruses
- RNA-mediated regulation in microbes
- Microbial genome evolution

Noam Stern-Ginossar
Profiling viral infection
- How viruses interface with and commandeer cellular pathways to control gene expression
- Uncover new aspects of virus-host interactions, as well as reveal new cell biology principles
- Deciphering the roles different viral elements are playing during infection


Talila Volk
The development and Cell biology of striated muscles
- The architecture and epigenetics of muscle nuclei
- Dynamics of myonuclei during muscle contraction
- Mechanotransduction and cell cycle progression in myofibers

Ernest Winocour

Elazar Zelzer
Musculoskeletal development, regeneration and pathology
- Our main goal is to understand the biological and biomechanical principles governing musculoskeletal development and function, maintenance and regeneration, as well as aging and pathology
- We investigate musculoskeletal assembly, that is, the creation of physical attachments between components of the system, focusing on the unique transitional tissue of the enthesis, the attachment site between tendon and bone
- We study the involvement of proprioceptive mechanosensors, which control muscle action, in musculoskeletal maintenance and disease, including scoliosis, developmental dysplasia of the hip (DDH) and fracture repair
- By using advanced imaging modalities, high-throughput computer vision algorithms and computational models for analyzing temporal series of 3D images, we study the mechanisms that sculpt the distinctive and intricate three-dimensional morphology of each bone
Professor Emeritus
-

Prof. Chaim Kahana
-

Prof. Ernest Winocour
-

Prof. Michel Revel
In Memorium
- Prof. Al Kaye
- Prof. Peter Lonai
- Prof. Yossi Aloni
- Prof. Leo Sachs
- Prof. Ben-Zion Shilo





























































































































































































































































