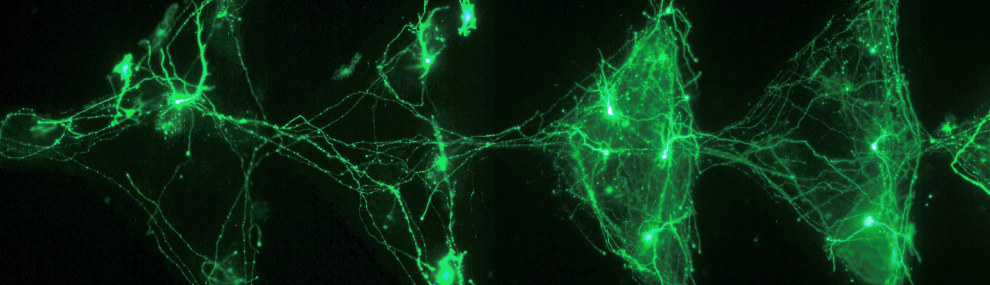
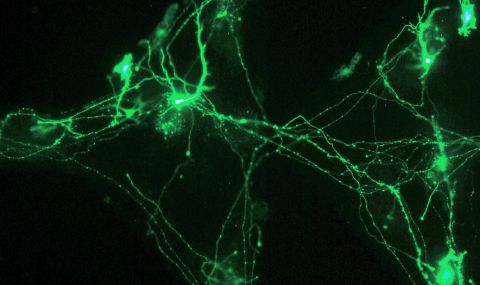
This work on Protein Arrays is done in collaboration with Dr. Ariel Lindner and the work on DNA arrays is done in collaboration with Dr. Shirely Horn Saban.
The aim of our project is to develop a "protein chip" and a "DNA chip", arrays of proteins or DNA, that will be able to detect proteins in concentrations that are not detectable in current methods and provide more quantitative and sensitive analysis of expression levels, protein interactions and protein circuits. Once such a "protein chip" or a "DNA chip" will be developed, different biological question that require the ability to detect very minute amount of molecules, and also requires the ability to give a quantitative description of these minute amounts, will be addressed.
We propose a method that reaches single molecule detection levels, gives high signal to noise ratios, and demonstrates a broad dynamic range, while retaining easy sample preparation and potential for high throughput abilities. Our method uses gold labeling and electron microscopy to probe DNA arrays, protein function arrays and protein detecting arrays (view cartoon, view example of gold labeled chip).
The reasons gold labeling and SEM scanning demonstrates such abilities are several:
- Noise is reduced significantly. Unlike fluorescence techniques where auto fluorescence of the glass and the buffers contribute to the noise, in our method false positive signals are only due to nonspecific binding of the gold conjugated protein probe to the slide. This is because only signal (back scattered electrons) coming from very heavy atoms such as gold can be detected by the SEM detectors.
- In small enough dilutions, where the molecules attached to the slide are in distances larger than the diameter of the gold probe, only one gold particle can attach per probed protein or ligand. This is because once a gold particle attached, via its conjugated protein, it occludes the protein on the surface from other gold probes. Since the SEM can detect single gold particles, this provides single molecule detection capabilities.
Detection abilities are limited in the lower limit only by false positive signals due to unspecific binding, and in the upper limit by the highest number of colloids able to pack closely in a given area. In other words, the upper detection limit can be controlled by the size of the gold colloids chosen as probes. Also, detection abilities are not constrained by instrumentation (unlike fluorescent methods where saturation of the photodiodes in light detectors can occur). In addition to the increased abilities in sensitivity and dynamic range there are several other advantages over other labeling methods such as fluorescence:
- There is no bleaching of the signal. This means that the sample can be stored and scanned repeatedly across a long period of time (i.e. in cases that inconsistencies were found in the analysis or new data has accumulated that requires new analysis of an old sample).
- The samples have a higher reproducibility rate since there is no dependence on the type of buffers and materials used, and there is a weak dependence on the "Hands" preparing the sample.
- It seems the gold enhances selectivity of the protein bound to it possibly because there are several proteins per gold which can increase the probability of binding. Another possibility is that due to the large size of the gold only high affinity and specific interactions survive the stringent washing conditions.
The potential of this method for high throughput is demonstrated in the semiconductor industry where SEMs are routinely used to scan wafers for microscopic defects and impurities.