Github
Some of the published works are made available through our GitHub account at https://github.com/WIS-MICC-CellObservatory.
WIS-NeuroMath - is a software tool for automated analysis and quantification of fluorescent microscopy images of Nerve cells, in both in vivo and in vitro preparations. This tool detects candidate neurites using a state-of-the-art edge detection algorithm proposed by Galun, Basri and Brandt at the department of computer science and applied mathematics. The method allows accurate detection of neurites in challenging images. WIS-NeuroMath was developed by Ofra Golani and Meirav Galun in collaboration with Prof. Michael Fainzilber and Dr. Ida Rishal from the department of biological Chemistry. Recently we are using it also to quantify angiogenesis.
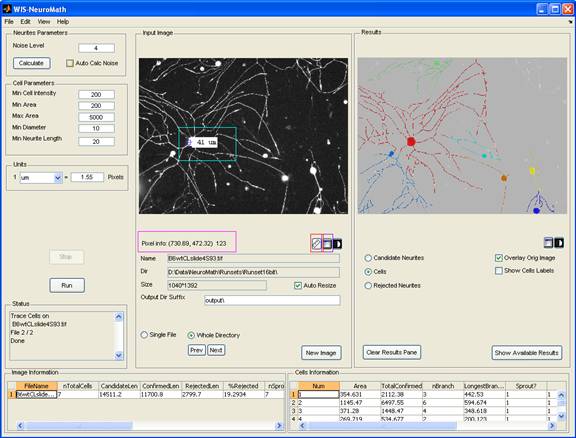
WIS-NeuroMath is based on accurate detection of candidate neurites using the algorithm described by Galun et al (2007). It first looks for edges in the image and then matches pairs of nearly parallel edges to find fibers, which constitute neurite-like structures. The method allows accurate detection of neurites in challenging images. Following neurite detection, different types of processing can be carried out depending on the desired application:
Cell Morphology of cultured neurons
Cell bodies are detected using the same image used for neurite detection. Candidate neurites are then traced to allow assignment to the relevant cell bodies. Neurite lengths, branching, cell body area and other parameters are then calculated and saved for each cell, while ignoring irrelevant items (neurites which are not attached to cell bodies, cell bodies that are too small or too big, etc.). Individual cell data are exported to Excel files, as well as image averages. The graphical user interface allows the user to modify detection thresholds and analysis parameters, to perform analyses on single files or whole directories, and to browse through the results.
Neurite Length Analysis on Sections
This mode provides a solution for images in which cell bodies are not present, for example longitudinal nerve sections that contain only neurites and non-neuronal cells. Neurites are detected and connected component analysis is applied to distinguish between neurite elements. A user-set threshold allows exclusion of very short neurite elements if desired, to reduce noise in certain experiments. The number of neurite elements, their average and median length are calculated as well as additional parameters such as the number of branches and the branching complexity.
Ganglion Explant Analysis
Ganglion explant cultures represent a particular challenge since neurites can be very profuse and dense close to the ganglion, and even a human eye cannot distinguish between them. Hence, in this mode the user manually defines an ellipse mask around the ganglion. The software then detects neurites and counts the number of neurites crossing that mask. Neurite numbers can be calculated at several offsets inside and outside the mask, to provide average and median numbers of crossing neurites. The user can set numbers of offset masks and the distance between them as desired.
Download
Please note that this download page is intended for research purposes only.
When you download the software you agree to the terms in the Software licence.
Download Software (zip file). The zip file includes WIS-NeuroMath executable together with few images that will enable you to test it. See documentation for installation instruction.
WIS-NeuroMath is a compiled Matlab application. To use it, you should download and install the proper version of Matlab Runtime Component (MCR)
on your computer: MCRInstaller for Win 32bit or MCRInstaller for Win 64bit.
News
Oct 10, 2011:
Version 3.4.8 is now available and supports many new features.
If you have any question please contact us.
Please credit WIS-NeuroMath by citing the following two papers
Citations:
- M. Galun, R. Basri and A. Brandt, "Multiscale edge detection and fiber enhancement using differences of oriented means", ICCV, 2007
- I. Rishal, O. Golani, M. Rajman, B. Costa, K. Ben-Yaakov, Z. Schoenmann, A. Yaron, R. Basri, M. Fainzilber and M. Galun, “WIS-NeuroMath enables versatile high throughput analyses of neuronal processes” Developmental Neurobiology, 2012.
We will appreciate if you would update us with any publication using the tool.
Publications Using Results Obtained With WIS-NeuroMath
- Naftelberg S., Abramovich Z., Gluska S, Yannai S, Joshi Y., Donyo M., Ben-Yaakov K., Gradus T., Zonszain J., Farhy C., Ashery-Padan R., Perlson E., Ast G. (2016), "Phosphatidylserine Ameliorates Neurodegenerative Symptoms and Enhances Axonal Transport in a Mouse Model of Familial Dysautonomia". PLOS Genetics. doi: 10.1371/jurnal.pgen.1006486
- Zuko A., Ouro-And A., Post H., Taggenbrock R.L.R.E., vanDijk R.E., Altelaar A.F.M., Heck A.J.R., Petrenko A.G., van der Zwaag B., Shimoda Y., Pasterkamp R.J., Burbach J.P.H.. (2016). "Association of Cell Adhesion Molecules Contactin-6 and Latrophilin-1 Regulates Neuronal Apoptosis". Frontiers in Molecular Neuroscience. doi 10.3389/fnmol.2016.00143
- Danelon V., Montroull L.E., Unsain N., Barker P.A., Masco D.H. (2016). "Calpain-dependent truncated form of TrkB-FL increases in neurodegenerative processes". Molecular and cellular Neuroscience. doi: 10.1016/j.mcn.2016.07.002
- Ben-Tov Perry R., Rishal, I., Doron-Mandel E., Kalinski A.L., Medzinhradsky K.F., Terenzio M. Alber S., Koley S., Lin A., Rozenbaum M., Yudin D., Sahoo P.K., Gomes C., Shinder V., Geraisy W., Huebner E.A., Woolf C.J., Yaron A., Burlingame A. L., Twiss J.L., Fainzilber M. (2016). "Nucleolin-Mediated RNA Localization Regulates Neuron Growth and Cycling Cell Size". Cell Reports. doi: 10.1016/j.celrep.2016.07.005
- Eckharter C., Junker N., Winter L., Fischer I., Fogli B., Kistner S., Pfaller K., Zheng B., Wiche G., Klimaschewski L., Schweigreiter R. (2015), "Schwann cell expressed Nogo-B modulates axonal branching of adult sensory neurons through the Nogo-B receptor NgBR". Frontiers in Cellular Neuroscience. doi: 10.3389/fncel.2015.00454
- Fannon, J., Tarmier, W. and Fulton, D. (2015), "Neuronal activity and AMPA-type glutamate receptor activation regulates the morphological development of oligodendrocyte precursor cells". Glia. doi: 10.1002/glia.22799
- Oguro-Ando A., Rosensweig C., Herman E., Nishimura Y., Werling D., Bill R., Berg J.M. Gao F., Coppola G., Abrahams B.S., Geschwind D.H. (2015), "Increased CYFIP1 dosage alters cellular and dendritic morphology and dysregulates mTOR". Molecular Pyschiatry. doi: 10.1038/mp.2014.124
- Marvaldi, L., Thongrong, S., Kozłowska, A., Irschick, R., Pritz, C. O., Bäumer, B., Ronchi, G., Geuna, S., Hausott, B. and Klimaschewski, L. (2014), "Enhanced axon outgrowth and improved long‐distance axon regeneration in sprouty2 deficient mice". Devel Neurobio. doi:10.1002/dneu.22224
- Painter M.W., Brosius-Lutz A., Cheng Y-C., Latremoliere A., Duong K., Miller C.M., Posada S., Cobos E.J, Zhang A.X., Wagers A.J., Havton L.A., Barres B., Omura T., Woolf C.J. (2014). "Diminished Schwann Cell repair Responses Underlie Age-Associated Impaired Axonal Regeneration". Neuron. doi: 10.1016/j.neuron.2014.06.016
- Rishal I., Kam N., Ben-Tov Perry R., Shinder V., Fisher E M.C. , Schiavo G. & Fainzilber M., 2012: "A Motor-Driven Mechanism for Cell-Length Sensing". Cell Reports, Volume 1, Issue 6, 608-616.
- Ben-Yaakov K., Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, Yudin D, Rishal I, Rother F, Bader M, Blesch A, Pilpel Y, Twiss JL & Fainzilber M., 2012: "Axonal transcription factors signal retrogradely in lesioned peripheral nerve". EMBO Journal 31, 1350 – 1363.
- C.H. Ma, G.J. Brenner, T. Omura, O.A. Samad, M. Costigan, P. Inquimbert, V. Niederkofler, R. Salie, H.Y. Lin, S. Arber, G. Coppola, C.J. Woolf, and T.A. Samad. (2011) The BMP co-receptor RGMb promotes while the endogenous BMP antagonist Noggin reduces neurite outgrowth and peripheral nerve regeneration by modulating BMP signaling. Journal of Neuroscience 31: 18391-18400.
WIS-PhagoTracker is a software application for quantitative analysis of high throughput cell migration assay. The cell migration assay [1] is based on a modified Phagokinetic tracks procedure, in which motile cells "leave their tracks" on a specialized surface. These tracks are visualized using a screening microscope.
WIS-PhagoTracker enables morphometric analysis of such tracks. It uses state of the art multiscale segmentation algorithm [2] for fine detection of tracks and cells boundaries. Following the segmentation step, it quantifies various morphometric parameters for each track, such as track area, perimeter, major and minor axis and solidity. All these measures are calculated for each track in each well of a well plate and saved for further statistical analysis.
![]()
WIS-PhagoTracker supports all the analysis phases starting from preprocessing, finding tracks of selected wells or a whole plate, through viewing the results and manually rejecting tracks to statistical analysis of the results. It also supports batch processing of several plates, and analysis of single image files. A user interface enables the user to modify the relevant parameters of the process, according to specific image's requirements. Results are exported into Excel readable files. It runs on Windows XP platforms. Users familiar with other Windows programs will find this software fairly straightforward to use.
WIS-PhagoTracker was developed by Ofra Golani, Meirav Galun and Suha Naffar Abu-Amara in the laboratories of Prof. Benny Geiger and Prof. Ronen Basri at the Department of Molecular Cell Biology and the Department of Computer Science and Applied Mathematics at the Weizmann Institute of Science.
The very accurate tracks detection which is the core of WIS-PhagoTracker is achieved by using multi-scale segmentation algorithm [2] developed by: Ronen Basri, Achi Brandt, Meirav Galun, Yoav Karnieli and Eitan Sharon, at the Department of Computer Science and Applied Mathematics at the Weizmann Institute and patented in [3].
Please credit WIS-PhagoTracker by citing the papers describing its underlying methods [1],[2]. We will appreciate it if you would update us with any publication using the tool.
References
- Naffar-Abu-Amara S, Shay T, Galun M, Cohen N, Isakoff SJ, Kam Z and Geiger B. Identification of novel pro-migratory, cancer-associated genes using quantitative, microscopy-based screening. PloS ONE. 2008 Jan 23;3(1): e1457.
- E.Sharon, M. Galun, D. Sharon, R.Basri and A. Brandt. Hierarchy and adaptivity in segmenting visual scenes. Nature, 442 (7104): 810-813 (2006).
- Achi Brandt, Eitan Sharon, and Ronen Basri. "Method and Apparatus for Data Clustering Including Segmentation and Boundary Detection". U.S. Patent and Trademark Office Application No. PCT/US01/43991, July, 2003, assigned to Yeda Research and Development Co., Ltd., Nov. 2000.
Download
Please note that this download page is intended only for academic research.
For a download password, please e-mail benny.geiger@weizmann.ac.il. When applying for a password, please make sure to include your name and institute name.
When you download the software you agree to the terms in the Software licence.
Download software (zip file). The zip file includes WIS-PhagoTracker executable together with few images that will enable you to test it. See documentation for installation instruction.
WIS-PhagoTracker is a compiled Matlab application. To use it, you should download and install the proper version of Matlab Runtime Component (MCR)
on your computer: MCRInstaller for Win 32bit or MCRInstaller for Win 64bit. In addition you should also install MCRInstaller 7.13. which is needed by the underlying segmentation program.
WIS-PhagoTracker can handle well montage and whole plates using the MRC (.r6d) format. If you have your montage in tif format, you can break the montage into single FOVs and pack them into MRC file using the Montage2MRC utility. If you have your files as single FOVs and you want to create a montage and pack them into MRC file you can use the PackWellMontage utility. Both utilities can be downloaded from MRC utilities. These are compiled Matlab applications and need the proper MCRInstaller (either Win 32bit or Win 64bit above).
If you have any question please contact the WIS-PhagoTracker team at ofra.golani@weizmann.ac.il.
News
- Monday, October 24, 2011
Updated version: 2.3.12, supports both Win 32bit and Win 64bit.
You need to install the proper MCRInstaller from this website, as the Matlab version was upgraded to 2010b. - Thursday, November 4, 2010
Updated version: 2.3.10.
You need to reinstall MCRInstaller from this website, as the Matlab version was upgraded to 2010a. - Wednesday, October 7, 2009
WIS-PhagoTracker was released to the public.
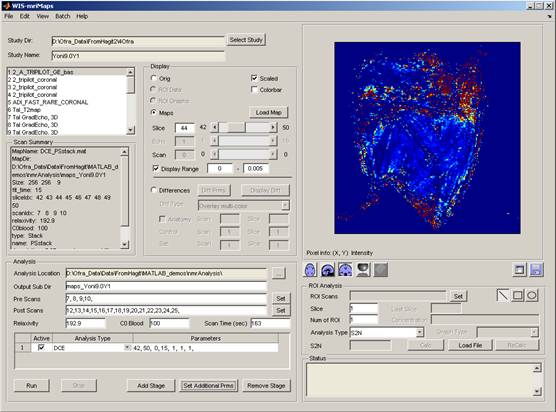
mriMaps - is a software application for analysis and display of MRI data. It was developed in collaboration with Prof. Michal Neeman, Dr. Hagit Dafni and Reut Avni from the department of Biological Regulation. Contact us for more information